DomPep--a general method for predicting modular domain-mediated protein-protein interactions
- PMID: 22003397
- PMCID: PMC3189207
- DOI: 10.1371/journal.pone.0025528
DomPep--a general method for predicting modular domain-mediated protein-protein interactions
Abstract
Protein-protein interactions (PPIs) are frequently mediated by the binding of a modular domain in one protein to a short, linear peptide motif in its partner. The advent of proteomic methods such as peptide and protein arrays has led to the accumulation of a wealth of interaction data for modular interaction domains. Although several computational programs have been developed to predict modular domain-mediated PPI events, they are often restricted to a given domain type. We describe DomPep, a method that can potentially be used to predict PPIs mediated by any modular domains. DomPep combines proteomic data with sequence information to achieve high accuracy and high coverage in PPI prediction. Proteomic binding data were employed to determine a simple yet novel parameter Ligand-Binding Similarity which, in turn, is used to calibrate Domain Sequence Identity and Position-Weighted-Matrix distance, two parameters that are used in constructing prediction models. Moreover, DomPep can be used to predict PPIs for both domains with experimental binding data and those without. Using the PDZ and SH2 domain families as test cases, we show that DomPep can predict PPIs with accuracies superior to existing methods. To evaluate DomPep as a discovery tool, we deployed DomPep to identify interactions mediated by three human PDZ domains. Subsequent in-solution binding assays validated the high accuracy of DomPep in predicting authentic PPIs at the proteome scale. Because DomPep makes use of only interaction data and the primary sequence of a domain, it can be readily expanded to include other types of modular domains.
Conflict of interest statement
Figures
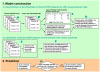


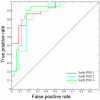

Similar articles
-
SH2 Ligand Prediction-Guidance for In-Silico Screening.Methods Mol Biol. 2017;1555:77-81. doi: 10.1007/978-1-4939-6762-9_5. Methods Mol Biol. 2017. PMID: 28092028
-
Predicting PDZ domain mediated protein interactions from structure.BMC Bioinformatics. 2013 Jan 21;14:27. doi: 10.1186/1471-2105-14-27. BMC Bioinformatics. 2013. PMID: 23336252 Free PMC article.
-
A regression framework incorporating quantitative and negative interaction data improves quantitative prediction of PDZ domain-peptide interaction from primary sequence.Bioinformatics. 2011 Feb 1;27(3):383-90. doi: 10.1093/bioinformatics/btq657. Epub 2010 Dec 2. Bioinformatics. 2011. PMID: 21127034 Free PMC article.
-
Domain-mediated protein interaction prediction: From genome to network.FEBS Lett. 2012 Aug 14;586(17):2751-63. doi: 10.1016/j.febslet.2012.04.027. Epub 2012 May 3. FEBS Lett. 2012. PMID: 22561014 Review.
-
Using peptide array to identify binding motifs and interaction networks for modular domains.Methods Mol Biol. 2009;570:67-76. doi: 10.1007/978-1-60327-394-7_3. Methods Mol Biol. 2009. PMID: 19649589 Review.
Cited by
-
The application of modular protein domains in proteomics.FEBS Lett. 2012 Aug 14;586(17):2586-96. doi: 10.1016/j.febslet.2012.04.019. Epub 2012 Apr 21. FEBS Lett. 2012. PMID: 22710164 Free PMC article. Review.
-
ShcA Adaptor Protein Promotes Nephrin Endocytosis and Is Upregulated in Proteinuric Nephropathies.J Am Soc Nephrol. 2018 Jan;29(1):92-103. doi: 10.1681/ASN.2017030285. Epub 2017 Oct 10. J Am Soc Nephrol. 2018. PMID: 29018139 Free PMC article.
-
Semi-supervised prediction of SH2-peptide interactions from imbalanced high-throughput data.PLoS One. 2013 May 17;8(5):e62732. doi: 10.1371/journal.pone.0062732. Print 2013. PLoS One. 2013. PMID: 23690949 Free PMC article.
-
Expression and Characterization of the Novel Gene fr47 during Freezing in the Wood Frog, Rana sylvatica.Biochem Res Int. 2015;2015:363912. doi: 10.1155/2015/363912. Epub 2015 May 26. Biochem Res Int. 2015. PMID: 26101667 Free PMC article.
-
Minimotif Miner 3.0: database expansion and significantly improved reduction of false-positive predictions from consensus sequences.Nucleic Acids Res. 2012 Jan;40(Database issue):D252-60. doi: 10.1093/nar/gkr1189. Epub 2011 Dec 6. Nucleic Acids Res. 2012. PMID: 22146221 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

