The two-pathway model of the biological catch-bond as a limit of the allosteric model
- PMID: 22004757
- PMCID: PMC3192973
- DOI: 10.1016/j.bpj.2011.09.005
The two-pathway model of the biological catch-bond as a limit of the allosteric model
Abstract
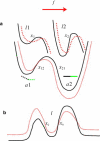
Catch-binding is a counterintuitive phenomenon in which the lifetime of a receptor/ligand bond increases when a force is applied to break the bond. Several mechanisms have been proposed to rationalize catch-binding. In the two-pathway model, the force drives the system away from its native dissociation pathway into an alternative pathway involving a higher energy barrier. Here, we analyze an allosteric model suggesting that a force applied to the complex alters the distribution of receptor conformations, and as a result, induces changes in the ligand-binding site. The model assumes explicitly that the allosteric transitions govern the properties of the ligand site. We demonstrate that the dynamics of the ligand is described by two relaxation times, one of which arises from the allosteric site. Therefore, we argue that one can characterize the allosteric transitions by studying the receptor/ligand binding. We show that the allosteric description reduces to the two-pathway model in the limit when the allosteric transitions are faster than the bond dissociation. The formal results are illustrated with two systems, P-selectin/PSGL-1 and FimH/mannose, subjected to both constant and time-dependent forces. The report advances our understanding of catch-binding by combining alternative physical models into a unified description and makes the problem more tractable for the bond mechanics community.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Theoretical aspects of the biological catch bond.Acc Chem Res. 2009 Jun 16;42(6):693-703. doi: 10.1021/ar800202z. Acc Chem Res. 2009. PMID: 19331389 Review.
-
Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond.Proc Natl Acad Sci U S A. 2004 Aug 3;101(31):11281-6. doi: 10.1073/pnas.0401870101. Epub 2004 Jul 26. Proc Natl Acad Sci U S A. 2004. PMID: 15277675 Free PMC article.
-
Allosteric coupling in the bacterial adhesive protein FimH.J Biol Chem. 2013 Aug 16;288(33):24128-39. doi: 10.1074/jbc.M113.461376. Epub 2013 Jul 2. J Biol Chem. 2013. PMID: 23821547 Free PMC article.
-
Catch-bond mechanism of the bacterial adhesin FimH.Nat Commun. 2016 Mar 7;7:10738. doi: 10.1038/ncomms10738. Nat Commun. 2016. PMID: 26948702 Free PMC article.
-
Catch-bond mechanism of force-enhanced adhesion: counterintuitive, elusive, but ... widespread?Cell Host Microbe. 2008 Oct 16;4(4):314-23. doi: 10.1016/j.chom.2008.09.005. Cell Host Microbe. 2008. PMID: 18854236 Free PMC article. Review.
Cited by
-
Cardiac desmosomal adhesion relies on ideal-, slip- and catch bonds.Sci Rep. 2024 Jan 31;14(1):2555. doi: 10.1038/s41598-024-52725-w. Sci Rep. 2024. PMID: 38297017 Free PMC article.
-
Molecular design of the γδT cell receptor ectodomain encodes biologically fit ligand recognition in the absence of mechanosensing.Proc Natl Acad Sci U S A. 2021 Jun 29;118(26):e2023050118. doi: 10.1073/pnas.2023050118. Proc Natl Acad Sci U S A. 2021. PMID: 34172580 Free PMC article.
-
Affinity Selection in Germinal Centers: Cautionary Tales and New Opportunities.Cells. 2021 Apr 28;10(5):1040. doi: 10.3390/cells10051040. Cells. 2021. PMID: 33924933 Free PMC article.
-
Dynamics of Mechanosensitive Nascent Adhesion Formation.Biophys J. 2019 Sep 17;117(6):1057-1073. doi: 10.1016/j.bpj.2019.08.004. Epub 2019 Aug 12. Biophys J. 2019. PMID: 31493858 Free PMC article.
-
Selectin catch-bonds mechanotransduce integrin activation and neutrophil arrest on inflamed endothelium under shear flow.Blood. 2017 Nov 9;130(19):2101-2110. doi: 10.1182/blood-2017-05-783027. Epub 2017 Aug 15. Blood. 2017. PMID: 28811304 Free PMC article. Clinical Trial.
References
-
- Thomas W.E., Trintchina E., Sokurenko E.V. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. - PubMed
-
- Marshall B.T., Long M., Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. - PubMed
-
- Sarangapani K.K., Yago T., Zhu C. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J. Biol. Chem. 2004;279:2291–2298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

