Allosteric regulation of phenylalanine hydroxylase
- PMID: 22005392
- PMCID: PMC3271142
- DOI: 10.1016/j.abb.2011.09.012
Allosteric regulation of phenylalanine hydroxylase
Abstract
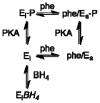
The liver enzyme phenylalanine hydroxylase is responsible for conversion of excess phenylalanine in the diet to tyrosine. Phenylalanine hydroxylase is activated by phenylalanine; this activation is inhibited by the physiological reducing substrate tetrahydrobiopterin. Phosphorylation of Ser16 lowers the concentration of phenylalanine for activation. This review discusses the present understanding of the molecular details of the allosteric regulation of the enzyme.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Fitzpatrick PF. Ann Rev Biochem. 1999;68:355–381. - PubMed
-
- Almas B, Le Bourdelles B, Flatmark T, Mallet J, Haavik J. Eur J Biochem. 1992;209:249–255. - PubMed
-
- Ramsey AJ, Fitzpatrick PF. Biochemistry. 1998;37:8980–8986. - PubMed
-
- McKinney J, Knappskog PM, Haavik J. J Neurochem. 2005;92:311–320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

