Design and development of a field-deployable single-molecule detector (SMD) for the analysis of molecular markers
- PMID: 22005669
- PMCID: PMC3278993
- DOI: 10.1039/c1an15658f
Design and development of a field-deployable single-molecule detector (SMD) for the analysis of molecular markers
Abstract
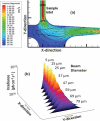
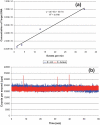
Single-molecule detection (SMD) has demonstrated some attractive benefits for many types of biomolecular analyses including enhanced processing speed by eliminating processing steps, elimination of ensemble averaging and single-molecule sensitivity. However, it's wide spread use has been hampered by the complex instrumentation required for its implementation when using fluorescence as the readout modality. We report herein a simple and compact fluorescence single-molecule instrument that is straightforward to operate and consisted of fiber optics directly coupled to a microfluidic device. The integrated fiber optics served as waveguides to deliver the laser excitation light to the sample and collecting the resulting emission, simplifying the optical requirements associated with traditional SMD instruments by eliminating the need for optical alignment and simplification of the optical train. Additionally, the use of a vertical cavity surface emitting laser and a single photon avalanche diode serving as the excitation source and photon transducer, respectively, as well as a field programmable gate array (FPGA) integrated into the processing electronics assisted in reducing the instrument footprint. This small footprint SMD platform was tested using fluorescent microspheres and single AlexaFluor 660 molecules to determine the optimal operating parameters and system performance. As a demonstration of the utility of this instrument for biomolecular analyses, molecular beacons (MBs) were designed to probe bacterial cells for the gene encoding Gram-positive species. The ability to monitor biomarkers using this simple and portable instrument will have a number of important applications, such as strain-specific detection of pathogenic bacteria or the molecular diagnosis of diseases requiring rapid turn-around-times directly at the point-of-use.
This journal is © The Royal Society of Chemistry 2012
Figures





References
-
- Gell C, Sabir T, Westwood J, Rashid A, Smith DAM, Harris SA, Stockley PG. J. Mol. Biol. 2008;384:264–278. - PubMed
-
- Haupts U, Rudiger M, Ashman S, Turconi S, Bingham R, Wharton C, Hutchinson J, Carey C, Moore KJ, Pope AJ. J. Biomol. Screening. 2003;8:19–33. - PubMed
-
- Hintersteiner M, Auer M. In: Fluorescence Methods and Applications: Spectroscopy, Imaging, and Probes. Wolfbeis OS, editor. vol. 1130. 2008. pp. 1–11.
-
- Kang SH, Kim YJ, Yeung ES. Anal. Bioanal. Chem. 2007;387:2663–2671. - PubMed
-
- Moore KJ, Turconi S, Ashman S, Ruediger M, Haupts U, Emerick V, Pope AJ. J. Biomol. Screening. 1999;4:335–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

