SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A)
- PMID: 22006005
- PMCID: PMC3256050
- DOI: 10.1128/AAC.05027-11
SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A)
Abstract
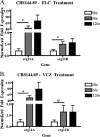
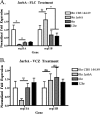
As triazole antifungal drug resistance during invasive Aspergillus fumigatus infection has become more prevalent, the need to understand mechanisms of resistance in A. fumigatus has increased. The presence of two erg11 (cyp51) genes in Aspergillus spp. is hypothesized to account for the inherent resistance of this mold to the triazole fluconazole (FLC). Recently, an A. fumigatus null mutant of a transcriptional regulator in the sterol regulatory element binding protein (SREBP) family, the ΔsrbA strain, was found to have increased susceptibility to FLC and voriconazole (VCZ). In this study, we examined the mechanism engendering the observed increase in A. fumigatus triazole susceptibility in the absence of SrbA. We observed a significant reduction in the erg11A transcript in the ΔsrbA strain in response to FLC and VCZ. Transcript levels of erg11B were also reduced but not to the extent of erg11A. Interestingly, erg11A transcript levels increased upon extended VCZ, but not FLC, exposure. Construction of an erg11A conditional expression strain in the ΔsrbA strain was able to restore erg11A transcript levels and, consequently, wild-type MICs to the triazole FLC. The VCZ MIC was also partially restored upon increased erg11A transcript levels; however, total ergosterol levels remained significantly reduced compared to those of the wild type. Induction of the erg11A conditional strain did not restore the hypoxia growth defect of the ΔsrbA strain. Taken together, our results demonstrate a critical role for SrbA-mediated regulation of ergosterol biosynthesis and triazole drug interactions in A. fumigatus that may have clinical importance.
Figures
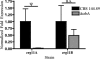
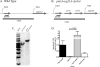


Similar articles
-
A secondary mechanism of action for triazole antifungals in Aspergillus fumigatus mediated by hmg1.Nat Commun. 2024 Apr 29;15(1):3642. doi: 10.1038/s41467-024-48029-2. Nat Commun. 2024. PMID: 38684680 Free PMC article.
-
SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus.PLoS Genet. 2011 Dec;7(12):e1002374. doi: 10.1371/journal.pgen.1002374. Epub 2011 Dec 1. PLoS Genet. 2011. PMID: 22144905 Free PMC article.
-
The CCAAT-binding complex mediates azole susceptibility of Aspergillus fumigatus by suppressing SrbA expression and cleavage.Microbiologyopen. 2021 Nov;10(6):e1249. doi: 10.1002/mbo3.1249. Microbiologyopen. 2021. PMID: 34964293 Free PMC article.
-
Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature.Antimicrob Agents Chemother. 2012 Jan;56(1):10-6. doi: 10.1128/AAC.05088-11. Epub 2011 Oct 17. Antimicrob Agents Chemother. 2012. PMID: 22005994 Free PMC article. Review.
-
Azole resistance in Aspergillus fumigatus- comprehensive review.Arch Microbiol. 2024 Jun 15;206(7):305. doi: 10.1007/s00203-024-04026-z. Arch Microbiol. 2024. PMID: 38878211 Review.
Cited by
-
Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter.BMC Genomics. 2012 Feb 6;13:62. doi: 10.1186/1471-2164-13-62. BMC Genomics. 2012. PMID: 22309491 Free PMC article.
-
An LaeA- and BrlA-Dependent Cellular Network Governs Tissue-Specific Secondary Metabolism in the Human Pathogen Aspergillus fumigatus.mSphere. 2018 Mar 14;3(2):e00050-18. doi: 10.1128/mSphere.00050-18. eCollection 2018 Mar-Apr. mSphere. 2018. PMID: 29564395 Free PMC article.
-
The Aspergillus fumigatus Damage Resistance Protein Family Coordinately Regulates Ergosterol Biosynthesis and Azole Susceptibility.mBio. 2016 Feb 23;7(1):e01919-15. doi: 10.1128/mBio.01919-15. mBio. 2016. PMID: 26908577 Free PMC article.
-
Coordination of hypoxia adaptation and iron homeostasis in human pathogenic fungi.Front Microbiol. 2012 Nov 6;3:381. doi: 10.3389/fmicb.2012.00381. eCollection 2012. Front Microbiol. 2012. PMID: 23133438 Free PMC article.
-
The C2H2-Type Transcription Factor ZfpA, Coordinately with CrzA, Affects Azole Susceptibility by Regulating the Multidrug Transporter Gene atrF in Aspergillus fumigatus.Microbiol Spectr. 2023 Aug 17;11(4):e0032523. doi: 10.1128/spectrum.00325-23. Epub 2023 Jun 15. Microbiol Spectr. 2023. PMID: 37318356 Free PMC article.
References
-
- Albarrag AM, et al. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 55:5113–5121 - PMC - PubMed
-
- Alcazar-Fuoli L, Mellado E, Cuenca-Estrella M, Sanglard D. 2011. Probing the role of point mutations in the cyp51A gene from Aspergillus fumigatus in the model yeast Saccharomyces cerevisiae. Med. Mycol. 49:276–284 - PubMed
-
- Alcazar-Fuoli L, et al. 2008. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73:339–347 - PubMed
-
- Alexander BD, Perfect JR. 1997. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs 54:657–678 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

