Identification of hopanoid biosynthesis genes involved in polymyxin resistance in Burkholderia multivorans
- PMID: 22006009
- PMCID: PMC3256039
- DOI: 10.1128/AAC.00602-11
Identification of hopanoid biosynthesis genes involved in polymyxin resistance in Burkholderia multivorans
Abstract
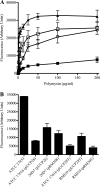
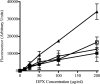
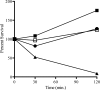
A major challenge to clinical therapy of Burkholderia cepacia complex (Bcc) pulmonary infections is their innate resistance to a broad range of antimicrobials, including polycationic agents such as aminoglycosides, polymyxins, and cationic peptides. To identify genetic loci associated with this phenotype, a transposon mutant library was constructed in B. multivorans ATCC 17616 and screened for increased susceptibility to polymyxin B. Compared to the parent strain, mutant 26D7 exhibited 8- and 16-fold increases in susceptibility to polymyxin B and colistin, respectively. Genetic analysis of mutant 26D7 indicated that the transposon inserted into open reading frame (ORF) Bmul_2133, part of a putative hopanoid biosynthesis gene cluster. A strain with a mutation in another ORF in this cluster, Bmul_2134, was constructed and named RMI19. Mutant RMI19 also had increased polymyxin susceptibility. Hopanoids are analogues of eukaryotic sterols involved in membrane stability and barrier function. Strains with mutations in Bmul_2133 and Bmul_2134 showed increased permeability to 1-N-phenylnaphthylamine in the presence of increasing concentrations of polymyxin, suggesting that the putative hopanoid biosynthesis genes are involved in stabilizing outer membrane permeability, contributing to polymyxin resistance. Results from a dansyl-polymyxin binding assay demonstrated that polymyxin B does not bind well to the parent or mutant strains, suggesting that Bmul_2133 and Bmul_2134 contribute to polymyxin B resistance by a mechanism that is independent of lipopolysaccharide (LPS) binding. Through this work, we propose a role for hopanoid biosynthesis as part of the multiple antimicrobial resistance phenotype in Bcc bacteria.
Figures
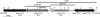
Similar articles
-
Fosmidomycin decreases membrane hopanoids and potentiates the effects of colistin on Burkholderia multivorans clinical isolates.Antimicrob Agents Chemother. 2014 Sep;58(9):5211-9. doi: 10.1128/AAC.02705-14. Epub 2014 Jun 23. Antimicrob Agents Chemother. 2014. PMID: 24957830 Free PMC article.
-
Surface changes and polymyxin interactions with a resistant strain of Klebsiella pneumoniae.Innate Immun. 2014 May;20(4):350-63. doi: 10.1177/1753425913493337. Epub 2013 Jul 25. Innate Immun. 2014. PMID: 23887184 Free PMC article.
-
Involvement of outer membrane of Pseudomonas cepacia in aminoglycoside and polymyxin resistance.Antimicrob Agents Chemother. 1986 Dec;30(6):923-6. doi: 10.1128/AAC.30.6.923. Antimicrob Agents Chemother. 1986. PMID: 3028253 Free PMC article.
-
Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia.Front Cell Infect Microbiol. 2011 Jul 22;1:6. doi: 10.3389/fcimb.2011.00006. eCollection 2011. Front Cell Infect Microbiol. 2011. PMID: 22919572 Free PMC article. Review.
-
Burkholderia cepacia complex infections: More complex than the bacterium name suggest.J Infect. 2018 Sep;77(3):166-170. doi: 10.1016/j.jinf.2018.07.006. Epub 2018 Jul 24. J Infect. 2018. PMID: 30012345 Review.
Cited by
-
Methylation at the C-2 position of hopanoids increases rigidity in native bacterial membranes.Elife. 2015 Jan 19;4:e05663. doi: 10.7554/eLife.05663. Elife. 2015. PMID: 25599566 Free PMC article.
-
Complete sequences of conjugal helper plasmids pRK2013 and pEVS104.MicroPubl Biol. 2023 Jul 13;2023:10.17912/micropub.biology.000882. doi: 10.17912/micropub.biology.000882. eCollection 2023. MicroPubl Biol. 2023. PMID: 37521139 Free PMC article.
-
Loss of O-Linked Protein Glycosylation in Burkholderia cenocepacia Impairs Biofilm Formation and Siderophore Activity and Alters Transcriptional Regulators.mSphere. 2019 Nov 13;4(6):e00660-19. doi: 10.1128/mSphere.00660-19. mSphere. 2019. PMID: 31722994 Free PMC article.
-
Pulmonary microbial spectrum of Burkholderia multivorans infection identified by metagenomic sequencing.Front Med (Lausanne). 2025 Jun 17;12:1577363. doi: 10.3389/fmed.2025.1577363. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40600049 Free PMC article.
-
Terpenoids and their biosynthesis in cyanobacteria.Life (Basel). 2015 Jan 21;5(1):269-93. doi: 10.3390/life5010269. Life (Basel). 2015. PMID: 25615610 Free PMC article. Review.
References
-
- Altas RM. 1993. Acid broth, p 52 In Parks LC. (ed), Handbook of microbiological media. CRC Press, Inc, Boca Raton, FL
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Ausubel F, et al. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY
-
- Bumford AA, Spilker T, LiPuma J. 2010. Epidemiology of Burkholderia infection in U.S. CF patients. International Burkholderia cepacia Working Group, Seattle, WA
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

