Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction
- PMID: 22006021
- PMCID: PMC3233019
- DOI: 10.1128/MCB.05694-11
Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction
Abstract
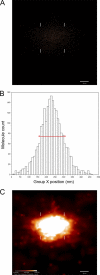
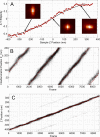
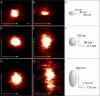
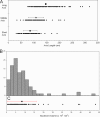
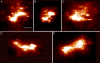
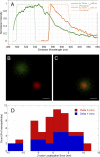
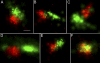
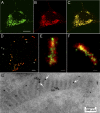
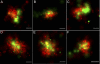
A fundamental objective in molecular biology is to understand how DNA is organized in concert with various proteins, RNA, and biological membranes. Mitochondria maintain and express their own DNA (mtDNA), which is arranged within structures called nucleoids. Their functions, dimensions, composition, and precise locations relative to other mitochondrial structures are poorly defined. Superresolution fluorescence microscopy techniques that exceed the previous limits of imaging within the small and highly compartmentalized mitochondria have been recently developed. We have improved and employed both two- and three-dimensional applications of photoactivated localization microscopy (PALM and iPALM, respectively) to visualize the core dimensions and relative locations of mitochondrial nucleoids at an unprecedented resolution. PALM reveals that nucleoids differ greatly in size and shape. Three-dimensional volumetric analysis indicates that, on average, the mtDNA within ellipsoidal nucleoids is extraordinarily condensed. Two-color PALM shows that the freely diffusible mitochondrial matrix protein is largely excluded from the nucleoid. In contrast, nucleoids are closely associated with the inner membrane and often appear to be wrapped around cristae or crista-like inner membrane invaginations. Determinations revealing high packing density, separation from the matrix, and tight association with the inner membrane underscore the role of mechanisms that regulate access to mtDNA and that remain largely unknown.
Figures


References
-
- Aquino D., et al. 2011. Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat. Methods 8:353–359 - PubMed
-
- Ashley N., Harris D., Poulton J. 2005. Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp. Cell Res. 303:432–446 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
