Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome
- PMID: 22006216
- PMCID: PMC3246206
- DOI: 10.1101/gr.124123.111
Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome
Erratum in
- Genome Res. 2012 Apr;22(4):810
Abstract
Analysis of diverse eukaryotes has revealed that recombination events cluster in discrete genomic locations known as hotspots. In humans, a zinc-finger protein, PRDM9, is believed to initiate recombination in >40% of hotspots by binding to a specific DNA sequence motif. However, the PRDM9 coding sequence is disrupted in the dog genome assembly, raising questions regarding the nature and control of recombination in dogs. By analyzing the sequences of PRDM9 orthologs in a number of dog breeds and several carnivores, we show here that this gene was inactivated early in canid evolution. We next use patterns of linkage disequilibrium using more than 170,000 SNP markers typed in almost 500 dogs to estimate the recombination rates in the dog genome using a coalescent-based approach. Broad-scale recombination rates show good correspondence with an existing linkage-based map. Significant variation in recombination rate is observed on the fine scale, and we are able to detect over 4000 recombination hotspots with high confidence. In contrast to human hotspots, 40% of canine hotspots are characterized by a distinct peak in GC content. A comparative genomic analysis indicates that these peaks are present also as weaker peaks in the panda, suggesting that the hotspots have been continually reinforced by accelerated and strongly GC biased nucleotide substitutions, consistent with the long-term action of biased gene conversion on the dog lineage. These results are consistent with the loss of PRDM9 in canids, resulting in a greater evolutionary stability of recombination hotspots. The genetic determinants of recombination hotspots in the dog genome may thus reflect a fundamental process of relevance to diverse animal species.
Figures
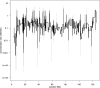


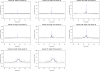


References
-
- Arnheim N, Calabrese P, Tiemann-Boege I 2007. Mammalian meiotic recombination hot spots. Annu Rev Genet 41: 369–399 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
