Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis
- PMID: 22006564
- PMCID: PMC3255689
- DOI: 10.1128/IAI.05496-11
Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis
Abstract
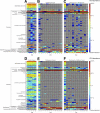
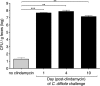
Antibiotic-induced changes in the intestinal microbiota predispose mammalian hosts to infection with antibiotic-resistant pathogens. Clostridium difficile is a Gram-positive intestinal pathogen that causes colitis and diarrhea in patients following antibiotic treatment. Clindamycin predisposes patients to C. difficile colitis. Here, we have used Roche-454 16S rRNA gene pyrosequencing to longitudinally characterize the intestinal microbiota of mice following clindamycin treatment in the presence or absence of C. difficile infection. We show that a single dose of clindamycin markedly reduces the diversity of the intestinal microbiota for at least 28 days, with an enduring loss of ca. 90% of normal microbial taxa from the cecum. Loss of microbial complexity results in dramatic sequential expansion and contraction of a subset of bacterial taxa that are minor contributors to the microbial consortium prior to antibiotic treatment. Inoculation of clindamycin-treated mice with C. difficile (VPI 10463) spores results in rapid development of diarrhea and colitis, with a 4- to 5-day period of profound weight loss and an associated 40 to 50% mortality rate. Recovering mice resolve diarrhea and regain weight but remain highly infected with toxin-producing vegetative C. difficile bacteria and, in comparison to the acute stage of infection, have persistent, albeit ameliorated cecal and colonic inflammation. The microbiota of "recovered" mice remains highly restricted, and mice remain susceptible to C. difficile infection at least 10 days following clindamycin, suggesting that resolution of diarrhea and weight gain may result from the activation of mucosal immune defenses.
Figures








References
-
- Bishara J, Peled N, Pitlik S, Samra Z. 2008. Mortality of patients with antibiotic-associated diarrhoea: the impact of Clostridium difficile. J. Hosp. Infect. 68: 308–314 - PubMed
-
- Chang JY, et al. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197: 435–438 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

