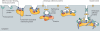Evolution: functional evolution of nuclear structure
- PMID: 22006947
- PMCID: PMC3198171
- DOI: 10.1083/jcb.201103171
Evolution: functional evolution of nuclear structure
Abstract
The evolution of the nucleus, the defining feature of eukaryotic cells, was long shrouded in speculation and mystery. There is now strong evidence that nuclear pore complexes (NPCs) and nuclear membranes coevolved with the endomembrane system, and that the last eukaryotic common ancestor (LECA) had fully functional NPCs. Recent studies have identified many components of the nuclear envelope in living Opisthokonts, the eukaryotic supergroup that includes fungi and metazoan animals. These components include diverse chromatin-binding membrane proteins, and membrane proteins with adhesive lumenal domains that may have contributed to the evolution of nuclear membrane architecture. Further discoveries about the nucleoskeleton suggest that the evolution of nuclear structure was tightly coupled to genome partitioning during mitosis.
Figures