Lactoferrin-endothelin-1 axis contributes to the development and invasiveness of triple-negative breast cancer phenotypes
- PMID: 22006997
- PMCID: PMC3617573
- DOI: 10.1158/0008-5472.CAN-11-1143
Lactoferrin-endothelin-1 axis contributes to the development and invasiveness of triple-negative breast cancer phenotypes
Abstract
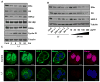
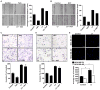
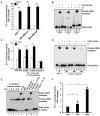
Triple-negative breast cancer (TNBC) is characterized by the lack of expression of estrogen receptor-α (ER-α), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER-2). However, pathways responsible for downregulation of therapeutic receptors, as well as subsequent aggressiveness, remain unknown. In this study, we discovered that lactoferrin (Lf) efficiently downregulates levels of ER-α, PR, and HER-2 in a proteasome-dependent manner in breast cancer cells, and it accounts for the loss of responsiveness to ER- or HER-2-targeted therapies. Furthermore, we found that lactoferrin increases migration and invasiveness of both non-TNBC and TNBC cell lines. We discovered that lactoferrin directly stimulates the transcription of endothelin-1 (ET-1), a secreted proinvasive polypeptide that acts through a specific receptor, ET(A)R, leading to secretion of the bioactive ET-1 peptide. Interestingly, a therapeutic ET-1 receptor-antagonist blocked lactoferrin-dependent motility and invasiveness of breast cancer cells. The physiologic significance of this newly discovered Lf-ET-1 axis in the manifestation of TNBC phenotypes is revealed by elevated plasma and tissue lactoferrin and ET-1 levels in patients with TNBC compared with those in ER(+) cases. These findings describe the first physiologically relevant polypeptide as a functional determinant in downregulating all three therapeutic receptors in breast cancer, which uses another secreted ET-1 system to confer invasiveness. Results presented in this article provide proof-of-principle evidence in support of the therapeutic effectiveness of ET-1 receptor antagonist to completely block the lactoferrin-induced motility and invasiveness of the TNBC as well as non-TNBC cells, and thus, open a remarkable opportunity to treat TNBC by targeting the Lf-ET-1 axis using an approved developmental drug.
Figures
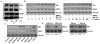


References
-
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;12:683–692. - PubMed
-
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;15(Pt 1):4429–4434. - PubMed
-
- Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;1:25–32. - PubMed
-
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;6871:530–536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

