A computational statistics approach for estimating the spatial range of morphogen gradients
- PMID: 22007136
- PMCID: PMC3201657
- DOI: 10.1242/dev.071571
A computational statistics approach for estimating the spatial range of morphogen gradients
Abstract
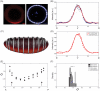
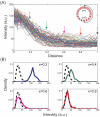
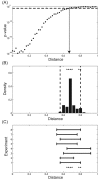
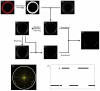
A crucial issue in studies of morphogen gradients relates to their range: the distance over which they can act as direct regulators of cell signaling, gene expression and cell differentiation. To address this, we present a straightforward statistical framework that can be used in multiple developmental systems. We illustrate the developed approach by providing a point estimate and confidence interval for the spatial range of the graded distribution of nuclear Dorsal, a transcription factor that controls the dorsoventral pattern of the Drosophila embryo.
Figures

References
-
- Affolter M., Basler K. (2007). The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 8, 663–674 - PubMed
-
- Ashe H. L., Briscoe J. (2006). The interpretation of morphogen gradients. Development 133, 385–394 - PubMed
-
- Bretzner L., Lindeberg T. (1998). Feature tracking with automatic selection of spatial scales. Computer Vision Image Understanding 71, 385–392
-
- Chopra V. S., Levine M. (2009). Combinatorial patterning mechanisms in the Drosophila embryo. Brief Funct. Genomic. Proteomic. 8, 243–249 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

