Kinetic analysis of the unique error signature of human DNA polymerase ν
- PMID: 22008035
- PMCID: PMC3235957
- DOI: 10.1021/bi201197p
Kinetic analysis of the unique error signature of human DNA polymerase ν
Abstract
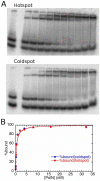
The fidelity of DNA synthesis by A-family DNA polymerases ranges from very accurate for bacterial, bacteriophage, and mitochondrial family members to very low for certain eukaryotic homologues. The latter include DNA polymerase ν (Pol ν) which, among all A-family polymerases, is uniquely prone to misincorporating dTTP opposite template G in a highly sequence-dependent manner. Here we present a kinetic analysis of this unusual error specificity, in four different sequence contexts and in comparison to Pol ν's more accurate A-family homologue, the Klenow fragment of Escherichia coli DNA polymerase I. The kinetic data strongly correlate with rates of stable misincorporation during gap-filling DNA synthesis. The lower fidelity of Pol ν compared to that of Klenow fragment can be attributed primarily to a much lower catalytic efficiency for correct dNTP incorporation, whereas both enzymes have similar kinetic parameters for G-dTTP misinsertion. The major contributor to sequence-dependent differences in Pol ν error rates is the reaction rate, k(pol). In the sequence context where fidelity is highest, k(pol) for correct G-dCTP incorporation by Pol ν is ~15-fold faster than k(pol) for G-dTTP misinsertion. However, in sequence contexts where the error rate is higher, k(pol) is the same for both correct and mismatched dNTPs, implying that the transition state does not provide additional discrimination against misinsertion. The results suggest that Pol ν may be fine-tuned to function when high enzyme activity is not a priority and may even be disadvantageous and that the relaxed active-site specificity toward the G-dTTP mispair may be associated with its cellular function(s).
© 2011 American Chemical Society
Figures


References
-
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. - PubMed
-
- Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. - PubMed
-
- Pang M, McConnell M, Fisher PA. The Drosophila mus 308 gene product, implicated in tolerance of DNA interstrand crosslinks, is a nuclear protein found in both ovaries and embryos. DNA Repair (Amst) 2005;4:971–982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

