Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference
- PMID: 22009198
- PMCID: PMC3243599
- DOI: 10.1038/emboj.2011.377
Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference
Abstract
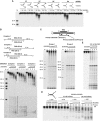
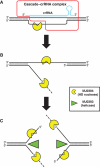
Clustered regularly interspaced short palindromic repeats (CRISPRs) and Cas proteins represent an adaptive microbial immunity system against viruses and plasmids. Cas3 proteins have been proposed to play a key role in the CRISPR mechanism through the direct cleavage of invasive DNA. Here, we show that the Cas3 HD domain protein MJ0384 from Methanocaldococcus jannaschii cleaves endonucleolytically and exonucleolytically (3'-5') single-stranded DNAs and RNAs, as well as 3'-flaps, splayed arms, and R-loops. The degradation of branched DNA substrates by MJ0384 is stimulated by the Cas3 helicase MJ0383 and ATP. The crystal structure of MJ0384 revealed the active site with two bound metal cations and together with site-directed mutagenesis suggested a catalytic mechanism. Our studies suggest that the Cas3 HD nucleases working together with the Cas3 helicases can completely degrade invasive DNAs through the combination of endo- and exonuclease activities.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66 (Pt 2): 213–221 - PMC - PubMed
-
- Alvarado J, Ghosh A, Janovitz T, Jauregui A, Hasson MS, Sanders DA (2006) Origin of exopolyphosphatase processivity: fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homolog. Structure 14: 1263–1272 - PubMed
-
- Aravind L, Koonin EV (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 23: 469–472 - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
-
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, Koonin EV, Edwards AM, Savchenko A, Yakunin AF (2008) A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem 283: 20361–20371 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

