The mitochondrial contact site complex, a determinant of mitochondrial architecture
- PMID: 22009199
- PMCID: PMC3230385
- DOI: 10.1038/emboj.2011.379
The mitochondrial contact site complex, a determinant of mitochondrial architecture
Abstract
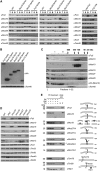
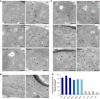
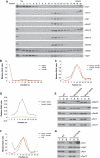
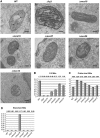
Mitochondria are organelles with a complex architecture. They are bounded by an envelope consisting of the outer membrane and the inner boundary membrane (IBM). Narrow crista junctions (CJs) link the IBM to the cristae. OMs and IBMs are firmly connected by contact sites (CS). The molecular nature of the CS remained unknown. Using quantitative high-resolution mass spectrometry we identified a novel complex, the mitochondrial contact site (MICOS) complex, formed by a set of mitochondrial membrane proteins that is essential for the formation of CS. MICOS is preferentially located at the CJs. Upon loss of one of the MICOS subunits, CJs disappear completely or are impaired, showing that CJs require the presence of CS to form a superstructure that links the IBM to the cristae. Loss of MICOS subunits results in loss of respiratory competence and altered inheritance of mitochondrial DNA.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture.Elife. 2015 Apr 28;4:e07739. doi: 10.7554/eLife.07739. Elife. 2015. PMID: 25918844 Free PMC article.
-
The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria.Mol Biol Cell. 2012 Jun;23(11):2143-55. doi: 10.1091/mbc.E11-10-0831. Epub 2012 Apr 11. Mol Biol Cell. 2012. PMID: 22496419 Free PMC article.
-
Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics.Biochim Biophys Acta Mol Cell Res. 2017 Apr;1864(4):737-746. doi: 10.1016/j.bbamcr.2016.05.020. Epub 2016 Sep 7. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 27614134 Review.
-
Coordination of cytochrome bc1 complex assembly at MICOS.EMBO Rep. 2025 Jan;26(2):353-384. doi: 10.1038/s44319-024-00336-x. Epub 2024 Dec 2. EMBO Rep. 2025. PMID: 39623166 Free PMC article.
-
Mitochondrial contact site and cristae organizing system.Curr Opin Cell Biol. 2016 Aug;41:33-42. doi: 10.1016/j.ceb.2016.03.013. Epub 2016 Apr 7. Curr Opin Cell Biol. 2016. PMID: 27062547 Review.
Cited by
-
MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria.J Cell Biol. 2016 Jun 6;213(5):525-34. doi: 10.1083/jcb.201602007. Epub 2016 May 30. J Cell Biol. 2016. PMID: 27241913 Free PMC article.
-
Tafazzins from Drosophila and mammalian cells assemble in large protein complexes with a short half-life.Mitochondrion. 2015 Mar;21:27-32. doi: 10.1016/j.mito.2015.01.002. Epub 2015 Jan 15. Mitochondrion. 2015. PMID: 25598000 Free PMC article.
-
Mitochondrial network morphology: building an integrative, geometrical view.BMC Biol. 2013 Jun 24;11:71. doi: 10.1186/1741-7007-11-71. BMC Biol. 2013. PMID: 23800141 Free PMC article. Review.
-
Biogenesis of mitochondrial β-barrel membrane proteins.FEBS Open Bio. 2024 Oct;14(10):1595-1609. doi: 10.1002/2211-5463.13905. Epub 2024 Sep 29. FEBS Open Bio. 2024. PMID: 39343721 Free PMC article. Review.
-
Mitochondrial Membrane Remodeling.Front Bioeng Biotechnol. 2022 Jan 4;9:786806. doi: 10.3389/fbioe.2021.786806. eCollection 2021. Front Bioeng Biotechnol. 2022. PMID: 35059386 Free PMC article. Review.
References
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Bayer ME (1991) Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J Struct Biol 107: 268–280 - PubMed
-
- Boldogh IR, Pon LA (2006) Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta 1763: 450–462 - PubMed
-
- Brdiczka DG, Zorov DB, Sheu SS (2006) Mitochondrial contact sites: their role in energy metabolism and apoptosis. Biochim Biophys Acta 1762: 148–163 - PubMed
-
- Chan D, Frank S, Rojo M (2006) Mitochondrial dynamics in cell life and death. Cell Death Differ 13: 680–684 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

