The chromatin-binding protein HMGN1 regulates the expression of methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice
- PMID: 22009741
- PMCID: PMC3234940
- DOI: 10.1074/jbc.M111.300541
The chromatin-binding protein HMGN1 regulates the expression of methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice
Abstract
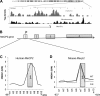
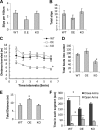
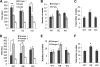
High mobility group N1 protein (HMGN1), a nucleosomal-binding protein that affects the structure and function of chromatin, is encoded by a gene located on chromosome 21 and is overexpressed in Down syndrome, one of the most prevalent genomic disorders. Misexpression of HMGN1 affects the cellular transcription profile; however, the biological function of this protein is still not fully understood. We report that HMGN1 modulates the expression of methyl CpG-binding protein 2 (MeCP2), a DNA-binding protein known to affect neurological functions including autism spectrum disorders, and whose alterations in HMGN1 levels affect the behavior of mice. Quantitative PCR and Western analyses of cell lines and brain tissues from mice that either overexpress or lack HMGN1 indicate that HMGN1 is a negative regulator of MeCP2 expression. Alterations in HMGN1 levels lead to changes in chromatin structure and histone modifications in the MeCP2 promoter. Behavior analyses by open field test, elevated plus maze, Reciprocal Social Interaction, and automated sociability test link changes in HMGN1 levels to abnormalities in activity and anxiety and to social deficits in mice. Targeted analysis of the Autism Genetic Resource Exchange genotype collection reveals a non-random distribution of genotypes within 500 kbp of HMGN1 in a region affecting its expression in families predisposed to autism spectrum disorders. Our results reveal that HMGN1 affects the behavior of mice and suggest that epigenetic changes resulting from altered HMGN1 levels could play a role in the etiology of neurodevelopmental disorders.
Figures

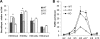

References
-
- Kugathasan S., Baldassano R. N., Bradfield J. P., Sleiman P. M., Imielinski M., Guthery S. L., Cucchiara S., Kim C. E., Frackelton E. C., Annaiah K., Glessner J. T., Santa E., Willson T., Eckert A. W., Bonkowski E., Shaner J. L., Smith R. M., Otieno F. G., Peterson N., Abrams D. J., Chiavacci R. M., Grundmeier R., Mamula P., Tomer G., Piccoli D. A., Monos D. S., Annese V., Denson L. A., Grant S. F., Hakonarson H. (2008) Nat. Genet. 40, 1211–1215 - PMC - PubMed
-
- Scherr R., Essers J., Hakonarson H., Kugathasan S. (2009) Dig. Dis. 27, 236–239 - PubMed
-
- Murata K., Degmetich S., Kinoshita M., Shimada E. (2009) Dev. Growth Differ. 51, 95–107 - PubMed
-
- Bustin M., Trieschmann L., Postnikov Y. V. (1995) Semin Cell Biol. 6, 247–255 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

