Profiling CCK-mediated pancreatic growth: the dynamic genetic program and the role of STATs as potential regulators
- PMID: 22010007
- PMCID: PMC3289124
- DOI: 10.1152/physiolgenomics.00255.2010
Profiling CCK-mediated pancreatic growth: the dynamic genetic program and the role of STATs as potential regulators
Abstract
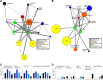
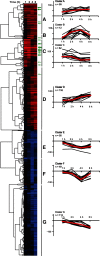
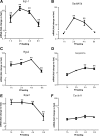
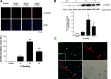
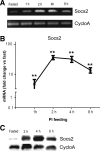
Feeding mice with protease inhibitor (PI) leads to increased endogenous cholecystokinin (CCK) release and results in pancreatic growth. This adaptive response requires calcineurin (CN)-NFAT and AKT-mTOR pathways, but the genes involved, the dynamics of their expression, and other regulatory pathways remain unknown. Here, we examined the early (1-8 h) transcriptional program that underlies pancreatic growth. We found 314 upregulated and 219 downregulated genes with diverse temporal and functional profiles. Several new identifications include the following: stress response genes Gdf15 and Txnip, metabolic mediators Pitpnc1 and Hmges2, as well as components of growth factor response Fgf21, Atf3, and Egr1. The genes fell into seven self-organizing clusters, each with a distinct pattern of expression; a representative gene within each of the upregulated clusters (Egr1, Gadd45b, Rgs2, and Serpinb1a) was validated by qRT-PCR. Genes up at any point throughout the time course and CN-dependent genes were subjected to further bioinformatics-based networking and promoter analysis, yielding STATs as potential transcriptional regulators. As shown by PCR, qPCR, and Western blots, the active phospho-form of STAT3 and the Jak-STAT feedback inhibitor Socs2 were both increased throughout early pancreatic growth. Moreover, immunohistochemistry showed a CCK-dependent and acinar cell-specific increase in nuclear localization of p-STAT3, with >75% nuclear occupancy in PI-fed mice vs. <0.1% in controls. Thus, the study identified novel genes likely to be important for CCK-driven pancreatic growth, characterized and biologically validated the dynamic pattern of their expression and investigated STAT-Socs signaling as a new player in this trophic response.
Figures


References
-
- Bragado MJ, Groblewski GE, Williams JA. Regulation of protein synthesis by cholecystokinin in rat pancreatic acini involves PHAS-I and the p70 S6 kinase pathway. Gastroenterology 115: 733–742, 1998. - PubMed
-
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942, 2005. - PubMed
-
- De Leon DD, Farzad C, Crutchlow MF, Brestelli J, Tobias J, Kaestner KH, Stoffers DA. Identification of transcriptional targets during pancreatic growth after partial pancreatectomy and exendin-4 treatment. Physiol Genomics 24: 133–143, 2006. - PubMed
-
- Elsasser H-P, Adler G, Kern HF. Replication and regeneration of the pancreas. In The Pancreas: Biology, Pathobiology, and Disease, edited by Vay Liang W. New York: Raven, 1993 p. 75–86.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

