Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis
- PMID: 22010036
- PMCID: PMC3229147
- DOI: 10.1105/tpc.111.087239
Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis
Abstract
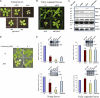
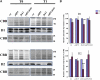
N-terminal methionine excision (NME) is the earliest modification affecting most proteins. All compartments in which protein synthesis occurs contain dedicated NME machinery. Developmental defects induced in Arabidopsis thaliana by NME inhibition are accompanied by increased proteolysis. Although increasing evidence supports a connection between NME and protein degradation, the identity of the proteases involved remains unknown. Here we report that chloroplastic NME (cNME) acts upstream of the FtsH protease complex. Developmental defects and higher sensitivity to photoinhibition associated with the ftsh2 mutation were abolished when cNME was inhibited. Moreover, the accumulation of D1 and D2 proteins of the photosystem II reaction center was always dependent on the prior action of cNME. Under standard light conditions, inhibition of chloroplast translation induced accumulation of correctly NME-processed D1 and D2 in a ftsh2 background, implying that the latter is involved in protein quality control, and that correctly NME-processed D1 and D2 are turned over primarily by the thylakoid FtsH protease complex. By contrast, inhibition of cNME compromises the specific N-terminal recognition of D1 and D2 by the FtsH complex, whereas the unprocessed forms are recognized by other proteases. Our results highlight the tight functional interplay between NME and the FtsH protease complex in the chloroplast.
Figures






References
-
- Adam Z., Clarke A.K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7: 451–456 - PubMed
-
- Adam Z., Rudella A., van Wijk K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9: 234–240 - PubMed
-
- Adir N., Ohad I. (1988). Structural properties of the D1 and surrounding photosystem II polypeptides as revealed by their interaction with cross-linking reagents. J. Biol. Chem. 263: 283–289 - PubMed
-
- Arfin S.M., Bradshaw R.A. (1988). Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry 27: 7979–7984 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

