Selective microRNA suppression in human thoracic aneurysms: relationship of miR-29a to aortic size and proteolytic induction
- PMID: 22010139
- PMCID: PMC3246193
- DOI: 10.1161/CIRCGENETICS.111.960419
Selective microRNA suppression in human thoracic aneurysms: relationship of miR-29a to aortic size and proteolytic induction
Abstract
Background: Increasing evidence points to a direct role for altered microRNA (miRNA or miR) expression levels in cardiovascular remodeling and disease progression. Although alterations in miR expression levels have been directly linked to cardiac hypertrophy, fibrosis, and remodeling, their role in regulating gene expression during thoracic aortic aneurysm (TAA) development has yet to be explored.
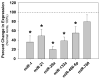
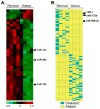
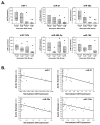
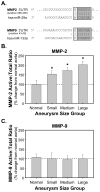
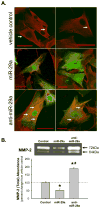
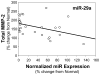
Methods and results: The present study examined miR expression levels in aortic tissue specimens collected from patients with ascending TAAs by quantitative real-time PCR, and observed decreased miR expression (miRs -1, -21, -29a, -133a, and -486) as compared with normal aortic specimens. A significant relationship between miR expression levels (miRs -1, -21, -29a, and -133a) and aortic diameter was identified; as aortic diameter increased, miR expression decreased. Through the use of a bioinformatics approach, members of the matrix metalloproteinase (MMP) family, proteins involved in TAA development, were examined for putative miR binding sites. MMP-2 and MMP-9 were identified as potential targets for miR-29a and miR-133a, respectively, and MMP-2 was subsequently verified as a miR-29a target in vitro. A significant inverse relationship between miR-29a and total MMP-2 was then identified in the clinical TAA specimens.
Conclusions: These findings demonstrate altered miR expression patterns in clinical TAA specimens, suggesting that the loss of specific miR expression may allow for the elaboration of specific MMPs capable of driving aortic remodeling during TAA development. Importantly, these data suggest that these miRs have biological and clinical relevance to the behavior of TAAs and may provide significant targets for therapeutic and diagnostic applications.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

