Theory and simulation of diffusion-influenced, stochastically gated ligand binding to buried sites
- PMID: 22010732
- PMCID: PMC3215080
- DOI: 10.1063/1.3645000
Theory and simulation of diffusion-influenced, stochastically gated ligand binding to buried sites
Abstract
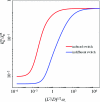
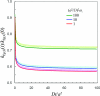
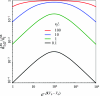
We consider the diffusion-influenced rate coefficient of ligand binding to a site located in a deep pocket on a protein; the binding pocket is flexible and can reorganize in response to ligand entrance. We extend to this flexible protein-ligand system a formalism developed previously [A. M. Berezhkovskii, A, Szabo, and H.-X. Zhou, J. Chem. Phys. 135, 075103 (2011)] for breaking the ligand-binding problem into an exterior problem and an interior problem. Conformational fluctuations of a bottleneck or a lid and the binding site are modeled as stochastic gating. We present analytical and Brownian dynamics simulation results for the case of a cylindrical pocket containing a binding site at the bottom. Induced switch, whereby the conformation of the protein adapts to the incoming ligand, leads to considerable rate enhancement.
© 2011 American Institute of Physics
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

