Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence
- PMID: 22010944
- PMCID: PMC3277930
- DOI: 10.1089/ars.2011.4341
Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence
Abstract
Significance: Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), can persist in a latent state for decades without causing overt disease. Since latent Mtb is refractory to current antimycobacterial drugs, the discovery and characterization of the biological mechanisms controlling the entry, maintenance, and emergence from latent infection is critical to the development of novel clinical therapies.
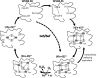
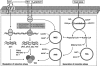
Recent advances: Recently, Mtb WhiB3, a member of the family of intracellular iron-sulfur (Fe-S) cluster proteins has emerged as a redox sensor and effector molecule controlling several aspects of Mtb virulence. WhiB3 was shown to contain a 4Fe-4S cluster that specifically reacts with important host gases (O(2) and NO), and exogenous and endogenous metabolic signals to maintain redox balance. Notably, the concept of reductive stress emerged from studies on WhiB3.
Critical issues: The detailed mechanism of how WhiB3 functions as an intracellular redox sensor is unknown. Sustaining Mtb redox balance is particularly important since the bacilli encounter a large number of redox stressors during infection, and because several antimycobacterial prodrugs are effective only upon bioreductive activation in the mycobacterial cytoplasm.
Future directions: How Mtb WhiB3 monitors its internal and external surroundings and modulates endogenous oxido-reductive pathways which in turn alter Mtb signal transduction, nucleic acid and protein synthesis, and enzymatic activation, is mostly unexplored. Modern expression, metabolomic and proteomic technologies should provide fresh insights into these yet unanswered questions.
Figures





References
-
- Agarwal N. Raghunand TR. Bishai WR. Regulation of the expression of whiB1 in Mycobacterium tuberculosis: Role of cAMP receptor protein. Microbiology. 2006;152:2749–2756. - PubMed
-
- Alam MS. Garg SK. Agrawal P. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: A [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. - PubMed
-
- Alam MS. Garg SK. Agrawal P. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 2009;276:76–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

