Structure and mechanisms of Escherichia coli aspartate transcarbamoylase
- PMID: 22011033
- PMCID: PMC3276696
- DOI: 10.1021/ar200166p
Structure and mechanisms of Escherichia coli aspartate transcarbamoylase
Abstract
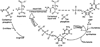
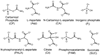
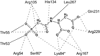
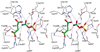
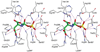
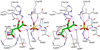
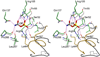
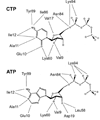
Enzymes catalyze a particular reaction in cells, but only a few control the rate of this reaction and the metabolic pathway that follows. One specific mechanism for such enzymatic control of a metabolic pathway involves molecular feedback, whereby a metabolite further down the pathway acts at a unique site on the control enzyme to alter its activity allosterically. This regulation may be positive or negative (or both), depending upon the particular system. Another method of enzymatic control involves the cooperative binding of the substrate, which allows a large change in enzyme activity to emanate from only a small change in substrate concentration. Allosteric regulation and homotropic cooperativity are often known to involve significant conformational changes in the structure of the protein. Escherichia coli aspartate transcarbamoylase (ATCase) is the textbook example of an enzyme that regulates a metabolic pathway, namely, pyrimidine nucleotide biosynthesis, by feedback control and by the cooperative binding of the substrate, L-aspartate. The catalytic and regulatory mechanisms of this enzyme have been extensively studied. A series of X-ray crystal structures of the enzyme in the presence and absence of substrates, products, and analogues have provided details, at the molecular level, of the conformational changes that the enzyme undergoes as it shifts between its low-activity, low-affinity form (T state) to its high-activity, high-affinity form (R state). These structural data provide insights into not only how this enzyme catalyzes the reaction between l-aspartate and carbamoyl phosphate to form N-carbamoyl-L-aspartate and inorganic phosphate, but also how the allosteric effectors modulate this activity. In this Account, we summarize studies on the structure of the enzyme and describe how these structural data provide insights into the catalytic and regulatory mechanisms of the enzyme. The ATCase-catalyzed reaction is regulated by nucleotide binding some 60 Å from the active site, inducing structural alterations that modulate catalytic activity. The delineation of the structure and function in this particular model system will help in understanding the molecular basis of cooperativity and allosteric regulation in other systems as well.
Figures



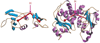


References
-
- Yates RA, Pardee AB. Control of pyrimidine biosynthesis in Escherichia coli. J. Biol. Chem. 1956;221:757–770. - PubMed
-
- Umbarger HE. Evidence for a Negative-Feedback Mechanism in the Biosynthesis of Isoleucine. Science. 1956;123:848–848. - PubMed
-
- Shepherdson M, Pardee AB. Production and Crystallization of Aspartate Transcarbamylase. J. Biol. Chem. 1960;235:3233–3237.
-
- Jones ME, Spector L, Lipmann F. Carbamyl phosphate. The carbamyl donor in enzymatic citrulline synthesis. J. Am. Chem. Soc. 1955;77:819–820.
-
- Lowenstein JM, Cohen PP. Studies on the Biosynthesis of carbamylaspartic acid. J. Biol. Chem. 1956;235:57–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

