Reactions of Nicotiana species to inoculation with monopartite and bipartite begomoviruses
- PMID: 22011413
- PMCID: PMC3213157
- DOI: 10.1186/1743-422X-8-475
Reactions of Nicotiana species to inoculation with monopartite and bipartite begomoviruses
Abstract
Background: Some Nicotiana species are widely used as experimental hosts for plant viruses. Nicotiana species differ in ploidy levels, chromosome numbers and have diverse geographical origins. Thus, these species are useful model systems to investigate virus-host interactions, co-evolution of pathogens and hosts and the effects of ploidy level on virus resistance/susceptibility.
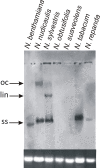
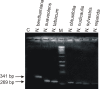
Results: Here we have studied the responses of seven Nicotiana species to inoculation with Cotton leaf curl Multan virus (CLCuMV), a monopartite begomovirus, and Tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus, both from the Indian subcontinent. All Nicotiana species supported the replication of both begomoviruses in inoculated leaves. However, only three Nicotiana species, namely N. benthamiana, N. tabacum and N. sylvestris showed symptoms when inoculated with ToLCNDV, while N. benthamiana was the only species that developed leaf curl symptoms when inoculated with CLCuMV. CLCuMV accumulated to detectable levels in N. tabacum, but plants remained asymptomatic. A previously identified mutation of RNA dependent RNA polymerase 1 was shown to be present only in N. benthamiana. The finding is in line with earlier results showing that the susceptibility of this species to a diverse range of plant viruses correlates with a defective RNA silencing-mediated host defense.
Conclusions: The results presented show that individual Nicotiana species respond differently to inoculation with begomoviruses. The inability of begomoviruses to systemically infect several Nicotiana species is likely due to inhibition of virus movement, rather than replication, and thus provides a novel model to study virus-host interactions in resistant/susceptible hosts.
Figures







Similar articles
-
Pepper leaf curl Lahore virus requires the DNA B component of Tomato leaf curl New Delhi virus to cause leaf curl symptoms.Virol J. 2010 Dec 13;7:367. doi: 10.1186/1743-422X-7-367. Virol J. 2010. PMID: 21144019 Free PMC article.
-
RNAi mediated broad-spectrum transgenic resistance in Nicotiana benthamiana to chilli-infecting begomoviruses.Plant Cell Rep. 2015 Aug;34(8):1389-99. doi: 10.1007/s00299-015-1795-8. Epub 2015 Apr 28. Plant Cell Rep. 2015. PMID: 25916177
-
Supervirulent pseudorecombination and asymmetric synergism between genomic components of two distinct species of begomovirus associated with severe tomato leaf curl disease in India.J Gen Virol. 2008 Mar;89(Pt 3):818-828. doi: 10.1099/vir.0.82873-0. J Gen Virol. 2008. PMID: 18272774
-
Tomato leaf curl New Delhi virus: a widespread bipartite begomovirus in the territory of monopartite begomoviruses.Mol Plant Pathol. 2017 Sep;18(7):901-911. doi: 10.1111/mpp.12481. Epub 2016 Oct 17. Mol Plant Pathol. 2017. PMID: 27553982 Free PMC article. Review.
-
Discovering host genes involved in the infection by the Tomato Yellow Leaf Curl Virus complex and in the establishment of resistance to the virus using Tobacco Rattle Virus-based post transcriptional gene silencing.Viruses. 2013 Mar 22;5(3):998-1022. doi: 10.3390/v5030998. Viruses. 2013. PMID: 23524390 Free PMC article. Review.
Cited by
-
Infectivity of Deinbollia mosaic virus, a novel weed-infecting begomovirus in East Africa.Arch Virol. 2017 Nov;162(11):3439-3445. doi: 10.1007/s00705-017-3495-x. Epub 2017 Aug 9. Arch Virol. 2017. PMID: 28791544 Free PMC article.
-
RNA-Seq Transcriptome Analysis Provides Candidate Genes for Resistance to Tomato Leaf Curl New Delhi Virus in Melon.Front Plant Sci. 2022 Jan 18;12:798858. doi: 10.3389/fpls.2021.798858. eCollection 2021. Front Plant Sci. 2022. PMID: 35116050 Free PMC article.
-
Directed Biosynthesis of New to Nature Alkaloids in a Heterologous Nicotiana benthamiana Expression Host.Front Plant Sci. 2022 Jun 22;13:919443. doi: 10.3389/fpls.2022.919443. eCollection 2022. Front Plant Sci. 2022. PMID: 35812900 Free PMC article.
-
CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana.PLoS Pathog. 2016 Jun 17;12(6):e1005668. doi: 10.1371/journal.ppat.1005668. eCollection 2016 Jun. PLoS Pathog. 2016. PMID: 27315204 Free PMC article.
References
-
- Stanley J, Bisaro DM, Briddon RW, Brown JK, Fauquet CM, Harrison BD, Rybicki EP, Stenger DC. In: Virus Taxonomy, VIIIth Report of the ICTV. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. London: Elsevier/Academic Press; 2005. Geminiviridae; pp. 301–326.
-
- Seal SE, van den Bosch F, Jeger MJ. Factors Influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit Rev Plant Sci. 2006;25:23–46. doi: 10.1080/07352680500365257. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

