Removing bias against membrane proteins in interaction networks
- PMID: 22011625
- PMCID: PMC3213014
- DOI: 10.1186/1752-0509-5-169
Removing bias against membrane proteins in interaction networks
Abstract
Background: Cellular interaction networks can be used to analyze the effects on cell signaling and other functional consequences of perturbations to cellular physiology. Thus, several methods have been used to reconstitute interaction networks from multiple published datasets. However, the structure and performance of these networks depends on both the quality and the unbiased nature of the original data. Due to the inherent bias against membrane proteins in protein-protein interaction (PPI) data, interaction networks can be compromised particularly if they are to be used in conjunction with drug screening efforts, since most drug-targets are membrane proteins.
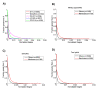
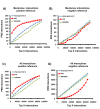
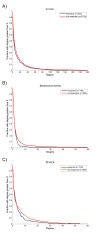
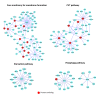
Results: To overcome the experimental bias against PPIs involving membrane-associated proteins we used a probabilistic approach based on a hypergeometric distribution followed by logistic regression to simultaneously optimize the weights of different sources of interaction data. The resulting less biased genome-scale network constructed for the budding yeast Saccharomyces cerevisiae revealed that the starvation pathway is a distinct subnetwork of autophagy and retrieved a more integrated network of unfolded protein response genes. We also observed that the centrality-lethality rule depends on the content of membrane proteins in networks.
Conclusions: We show here that the bias against membrane proteins can and should be corrected in order to have a better representation of the interactions and topological properties of protein interaction networks.
Figures





Similar articles
-
Functional centrality: detecting lethality of proteins in protein interaction networks.Genome Inform. 2007;19:166-77. Genome Inform. 2007. PMID: 18546514
-
t-LSE: a novel robust geometric approach for modeling protein-protein interaction networks.PLoS One. 2013;8(4):e58368. doi: 10.1371/journal.pone.0058368. Epub 2013 Apr 1. PLoS One. 2013. PMID: 23560036 Free PMC article.
-
Integrating transcriptional and protein interaction networks to prioritize condition-specific master regulators.BMC Syst Biol. 2015 Nov 14;9:80. doi: 10.1186/s12918-015-0228-1. BMC Syst Biol. 2015. PMID: 26576632 Free PMC article.
-
Integrating genetic and protein-protein interaction networks maps a functional wiring diagram of a cell.Curr Opin Microbiol. 2018 Oct;45:170-179. doi: 10.1016/j.mib.2018.06.004. Epub 2018 Jul 28. Curr Opin Microbiol. 2018. PMID: 30059827 Free PMC article. Review.
-
Zooming in on yeast osmoadaptation.Adv Exp Med Biol. 2012;736:293-310. doi: 10.1007/978-1-4419-7210-1_17. Adv Exp Med Biol. 2012. PMID: 22161336 Review.
Cited by
-
HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks.Nucleic Acids Res. 2017 Jan 4;45(D1):D408-D414. doi: 10.1093/nar/gkw985. Epub 2016 Oct 24. Nucleic Acids Res. 2017. PMID: 27794551 Free PMC article.
-
Geometric characterisation of disease modules.Appl Netw Sci. 2018;3(1):10. doi: 10.1007/s41109-018-0066-3. Epub 2018 Jun 18. Appl Netw Sci. 2018. PMID: 30839777 Free PMC article.
-
Evolutionary Influenced Interaction Pattern as Indicator for the Investigation of Natural Variants Causing Nephrogenic Diabetes Insipidus.Comput Math Methods Med. 2015;2015:641393. doi: 10.1155/2015/641393. Epub 2015 May 28. Comput Math Methods Med. 2015. PMID: 26180540 Free PMC article.
-
A comprehensive strategy to identify stoichiometric membrane protein interactomes.Cell Logist. 2012 Oct 1;2(4):189-196. doi: 10.4161/cl.22717. Cell Logist. 2012. PMID: 23676845 Free PMC article.
-
Neurodevelopmental disorders: mechanisms and boundary definitions from genomes, interactomes and proteomes.Transl Psychiatry. 2013 Dec 3;3(12):e329. doi: 10.1038/tp.2013.108. Transl Psychiatry. 2013. PMID: 24301647 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

