The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations
- PMID: 22012064
- PMCID: PMC3242717
- DOI: 10.1182/blood-2010-11-317909
The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations
Abstract
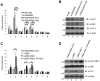
The mixed-lineage leukemia (MLL) H3K4 methyltransferase protein, and the heterodimeric RUNX1/CBFβ transcription factor complex, are critical for definitive and adult hematopoiesis, and both are frequently targeted in human acute leukemia. We identified a physical and functional interaction between RUNX1 (AML1) and MLL and show that both are required to maintain the histone lysine 4 trimethyl mark (H3K4me3) at 2 critical regulatory regions of the AML1 target gene PU.1. Similar to CBFβ, we show that MLL binds to AML1 abrogating its proteasome-dependent degradation. Furthermore, a subset of previously uncharacterized frame-shift and missense mutations at the N terminus of AML1, found in MDS and AML patients, impairs its interaction with MLL, resulting in loss of the H3K4me3 mark within PU.1 regulatory regions, and decreased PU.1 expression. The interaction between MLL and AML1 provides a mechanism for the sequence-specific binding of MLL to DNA, and identifies RUNX1 target genes as potential effectors of MLL function.
Figures

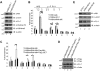


References
-
- Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5(9-10):1282–1297. - PubMed
-
- Yoshida H, Kitabayashi I. Chromatin regulation by AML1 complex. Int J Hematol. 2008;87(1):19–24. - PubMed
-
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278(5340):1059–1064. - PubMed
-
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691–700. - PubMed
-
- Gu Y, Nakamura T, Alder H, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71(4):701–708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

