Transcriptomic analyses of murine resolution-phase macrophages
- PMID: 22012065
- PMCID: PMC5362087
- DOI: 10.1182/blood-2011-04-345330
Transcriptomic analyses of murine resolution-phase macrophages
Abstract
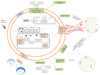
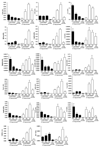
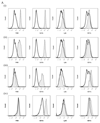
Macrophages are either classically (M1) or alternatively-activated (M2). Whereas this nomenclature was generated from monocyte-derived macrophages treated in vitro with defined cytokine stimuli, the phenotype of in vivo-derived macrophages is less understood. We completed Affymetrix-based transcriptomic analysis of macrophages from the resolution phase of a zymosan-induced peritonitis. Compared with macrophages from hyperinflamed mice possessing a pro-inflammatory nature as well as naive macrophages from the uninflamed peritoneum, resolution-phase macrophages (rM) are similar to monocyte-derived dendritic cells (DCs), being CD209a positive but lacking CD11c. They are enriched for antigen processing/presentation (MHC class II [H2-Eb1, H2-Ab1, H2-Ob, H2-Aa], CD74, CD86), secrete T- and B-lymphocyte chemokines (Xcl1, Ccl5, Cxcl13) as well as factors that enhance macrophage/DC development, and promote DC/T cell synapse formation (Clec2i, Tnfsf4, Clcf1). rM are also enriched for cell cycle/proliferation genes as well as Alox15, Timd4, and Tgfb2, key systems in the termination of leukocyte trafficking and clearance of inflammatory cells. Finally, comparison with in vitro-derived M1/M2 shows that rM are neither classically nor alternatively activated but possess aspects of both definitions consistent with an immune regulatory phenotype. We propose that macrophages in situ cannot be rigidly categorized as they can express many shades of the inflammatory spectrum determined by tissue, stimulus, and phase of inflammation.
Figures




















References
-
- Bowdish DM, Loffredo MS, Mukhopadhyay S, Mantovani A, Gordon S. Macrophage receptors implicated in the "adaptive" form of innate immunity. Microbes Infect. 2007;9(14-15):1680–1687. - PubMed
-
- Savill J. Phagocyte recognition of apoptotic cells. Biochem Soc Trans. 1996;24(4):1065–1069. - PubMed
-
- van Furth R. Mycobacteria and macrophage activation. Res Microbiol. 1990;141(2):256–261. - PubMed
-
- Rosenthal AS, Lipsky PE, Shevach EM. Macrophage-lymphocyte interaction and antigen recognition. Fed Proc. 1975;34(8):1743–1748. - PubMed
-
- Shapiro SD. Diverse roles of macrophage matrix metalloproteinases in tissue destruction and tumor growth. Thromb Haemost. 1999;82(2):846–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

