A systems view of genetics in chronic kidney disease
- PMID: 22012128
- PMCID: PMC4224965
- DOI: 10.1038/ki.2011.359
A systems view of genetics in chronic kidney disease
Abstract
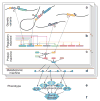
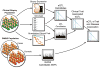
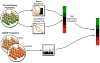
A tight interplay of genetic predisposition and environmental factors define the onset and the rate of progression of chronic renal disease. We are seeing a rapid expansion of information about genetic loci associated with kidney function and complex renal disease. However, discovering the functional links that bridge the gap from genetic risk loci to disease phenotype is one of the main challenges ahead. Risk loci are currently assigned to a putative context using the functional annotation of the closest genes via a guilt-by-proximity approach. These approaches can be extended by strategies integrating genetic risk loci with kidney-specific, genome-wide gene expression. Risk loci-associated transcripts can be assigned a putative disease-specific function using gene expression coregulation networks. Ultimately, genotype-phenotype dependencies postulated from these associative approaches in humans need to be tested via genetic modification in model organisms. In this review, we survey strategies that employ human tissue-specific expression and the use of model organisms to identify and validate the functional relationship between genotype and phenotype in renal disease. Strategies to unravel how genetic risk and environmental factors orchestrate renal disease manifestation can be the first steps toward a more integrated, holistic approach urgently needed for chronic renal diseases.
Conflict of interest statement
All authors have no competing interests.
Figures

References
-
- Ferguson R, Grim CE, Opgenorth TJ. A familial risk of chronic renal failure among blacks on dialysis? J Clin Epidemiol. 1988;41:1189–1196. - PubMed
-
- Ng DPK, Tai BC, Koh D, et al. Angiotensin-I converting enzyme insertion/deletion polymorphism and its association with diabetic nephropathy: a meta-analysis of studies reported between 1994 and 2004 and comprising 14,727 subjects. Diabetologia. 2005;48:1008–1016. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

