Signals for pre-mRNA cleavage and polyadenylation
- PMID: 22012871
- PMCID: PMC4451228
- DOI: 10.1002/wrna.116
Signals for pre-mRNA cleavage and polyadenylation
Abstract
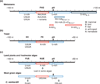
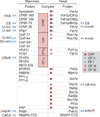
Pre-mRNA cleavage and polyadenylation is an essential step for 3' end formation of almost all protein-coding transcripts in eukaryotes. The reaction, involving cleavage of nascent mRNA followed by addition of a polyadenylate or poly(A) tail, is controlled by cis-acting elements in the pre-mRNA surrounding the cleavage site. Experimental and bioinformatic studies in the past three decades have elucidated conserved and divergent elements across eukaryotes, from yeast to human. Here we review histories and current models of these elements in a broad range of species.
Copyright © 2011 John Wiley & Sons, Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

