A framework to select clinically relevant cancer cell lines for investigation by establishing their molecular similarity with primary human cancers
- PMID: 22012889
- PMCID: PMC3242880
- DOI: 10.1158/0008-5472.CAN-11-2427
A framework to select clinically relevant cancer cell lines for investigation by establishing their molecular similarity with primary human cancers
Abstract
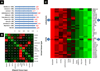
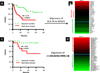
Experimental work on human cancer cell lines often does not translate to the clinic. We posit that this is because some cells undergo changes in vitro that no longer make them representative of human tumors. Here, we describe a novel alignment method named Spearman's rank correlation classification method (SRCCM) that measures similarity between cancer cell lines and human tumors via gene expression profiles, for the purpose of selecting lines that are biologically relevant. To show utility, we used SRCCM to assess similarity of 36 bladder cancer lines with 10 epithelial human tumor types (N = 1,630 samples) and with bladder tumors of different stages and grades (N = 144 samples). Although 34 of 36 lines aligned to bladder tumors rather than other histologies, only 16 of 28 had SRCCM assigned grades identical to that of their original source tumors. To evaluate the clinical relevance of this approach, we show that gene expression profiles of aligned cell lines stratify survival in an independent cohort of 87 bladder patients (HR = 3.41, log-rank P = 0.0077) whereas unaligned cell lines using original tumor grades did not. We repeated this process on 22 colorectal cell lines and found that gene expression profiles of 17 lines aligning to colorectal tumors and selected based on their similarity with 55 human tumors stratified survival in an independent cohort of 177 colorectal cancer patients (HR = 2.35, log-rank P = 0.0019). By selecting cell lines that reflect human tumors, our technique promises to improve the clinical translation of laboratory investigations in cancer.
Conflict of interest statement
Figures




References
-
- Inoue K, Slaton JW, Kim SJ, et al. Interleukin 8 expression regulates tumorigenicity and metastasis in human bladder cancer. Cancer Res. 2000;60:2290–2299. - PubMed
-
- Saitoh H, Mori K, Kudoh S, Itoh H, Takahashi N, Suzuki T. BCG effects on telomerase activity in bladder cancer cell lines. Int J Clin Oncol. 2002;7:165–170. - PubMed
-
- Zhang YB, Niu HT, Chang JW, Dong GL, Ma XB. EZH2 silencing by RNA interference inhibits proliferation in bladder cancer cell lines. Eur J Cancer Care (Engl) 2011;20:106–112. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

