The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H
- PMID: 22013041
- PMCID: PMC3255910
- DOI: 10.1128/JVI.06082-11
The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H
Abstract
The APOBEC3 family comprises seven cytidine deaminases (APOBEC3A [A3A] to A3H), which are expressed to various degrees in HIV-1 susceptible cells. The HIV-1 Vif protein counteracts APOBEC3 restriction by mediating its degradation by the proteasome. We hypothesized that Vif proteins from various HIV-1 subtypes differ in their abilities to counteract different APOBEC3 proteins. Seventeen Vif alleles from seven HIV-1 subtypes were tested for their abilities to degrade and counteract A3G, A3F, and A3H haplotype II (hapII). We show that most Vif alleles neutralize A3G and A3F efficiently but display differences with respect to the inhibition of A3H hapII. The majority of non-subtype B Vif alleles tested presented some activity against A3H hapII, with two subtype F Vif variants being highly effective in counteracting A3H hapII. The residues required for activity were mapped to two residues in the amino-terminal region of Vif (positions 39F and 48H). Coimmunoprecipitations showed that these two amino acids were necessary for association of Vif with A3H hapII. These findings suggest that the A3H hapII binding site in Vif is distinct from the regions important for A3G and A3F recognition and that it requires specific amino acids at positions 39 and 48. The differential Vif activity spectra, especially against A3H hapII, suggest adaptation to APOBEC3 repertoires representative of different human ancestries. Phenotypic assessment of anti-APOBEC3 activity of Vif variants against several cytidine deaminases will help reveal the requirement for successful replication in vivo and ultimately point to interventions targeting the Vif-APOBEC3 interface.
Figures

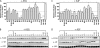
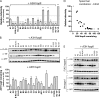




References
-
- Bell CM, et al. 2007. Molecular characterization of the HIV type 1 subtype C accessory genes vif, vpr, and vpu. AIDS Res. Hum. Retroviruses 23:322–330 - PubMed
-
- Conticello SG, Harris RS, Neuberger MS. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009–2013 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

