Small but sufficient: the Rhodococcus phage RRH1 has the smallest known Siphoviridae genome at 14.2 kilobases
- PMID: 22013058
- PMCID: PMC3255915
- DOI: 10.1128/JVI.05460-11
Small but sufficient: the Rhodococcus phage RRH1 has the smallest known Siphoviridae genome at 14.2 kilobases
Abstract
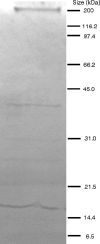
Bacteriophages are considered to be the most abundant biological entities on the planet. The Siphoviridae are the most commonly encountered tailed phages and contain double-stranded DNA with an average genome size of ∼50 kb. This paper describes the isolation from four different activated sludge plants of the phage RRH1, which is polyvalent, lysing five Rhodococcus species. It has a capsid diameter of only ∼43 nm. Whole-genome sequencing of RRH1 revealed a novel circularly permuted DNA sequence (14,270 bp) carrying 20 putative open reading frames. The genome has a modular arrangement, as reported for those of most Siphoviridae phages, but appears to encode only structural proteins and carry a single lysis gene. All genes are transcribed in the same direction. RRH1 has the smallest genome yet of any described functional Siphoviridae phage. We demonstrate that lytic phage can be recovered from transforming naked DNA into its host bacterium, thus making it a potentially useful model for studying gene function in phages.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources

