Structural basis for the substrate specificity of a novel β-N-acetylhexosaminidase StrH protein from Streptococcus pneumoniae R6
- PMID: 22013074
- PMCID: PMC3234876
- DOI: 10.1074/jbc.M111.256578
Structural basis for the substrate specificity of a novel β-N-acetylhexosaminidase StrH protein from Streptococcus pneumoniae R6
Abstract
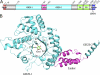
The β-N-acetylhexosaminidase (EC 3.2.1.52) from glycoside hydrolase family 20 (GH20) catalyzes the hydrolysis of the β-N-acetylglucosamine (NAG) group from the nonreducing end of various glycoconjugates. The putative surface-exposed N-acetylhexosaminidase StrH/Spr0057 from Streptococcus pneumoniae R6 was proved to contribute to the virulence by removal of β(1,2)-linked NAG on host defense molecules following the cleavage of sialic acid and galactose by neuraminidase and β-galactosidase, respectively. StrH is the only reported GH20 enzyme that contains a tandem repeat of two 53% sequence-identical catalytic domains (designated as GH20-1 and GH20-2, respectively). Here, we present the 2.1 Å crystal structure of the N-terminal domain of StrH (residues Glu-175 to Lys-642) complexed with NAG. It adopts an overall structure similar to other GH20 enzymes: a (β/α)(8) TIM barrel with the active site residing at the center of the β-barrel convex side. The kinetic investigation using 4-nitrophenyl N-acetyl-β-d-glucosaminide as the substrate demonstrated that GH20-1 had an enzymatic activity (k(cat)/K(m)) of one-fourth compared with GH20-2. The lower activity of GH20-1 could be attributed to the substitution of active site Cys-469 of GH20-1 to the counterpart Tyr-903 of GH20-2. A complex model of NAGβ(1,2)Man at the active site of GH20-1 combined with activity assays of the corresponding site-directed mutants characterized two key residues Trp-443 and Tyr-482 at subsite +1 of GH20-1 (Trp-876 and Tyr-914 of GH20-2) that might determine the β(1,2) substrate specificity. Taken together, these findings shed light on the mechanism of catalytic specificity toward the β(1,2)-linked β-N-acetylglucosides.
Figures

References
-
- García-Suárez Mdel M., Vázquez F., Méndez F. J. (2006) Enferm. Infecc. Microbiol. Clin. 24, 512–517 - PubMed
-
- Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Science 293, 498–506 - PubMed
-
- King S. J., Hippe K. R., Weiser J. N. (2006) Mol. Microbiol. 59, 961–974 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

