Validation of kinetics similarity in qPCR
- PMID: 22013160
- PMCID: PMC3287174
- DOI: 10.1093/nar/gkr778
Validation of kinetics similarity in qPCR
Abstract
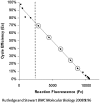
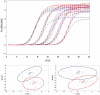
Quantitative real-time PCR (qPCR) is the method of choice for specific and sensitive quantification of nucleic acids. However, data validation is still a major issue, partially due to the complex effect of PCR inhibition on the results. If undetected PCR inhibition may severely impair the accuracy and sensitivity of results. PCR inhibition is addressed by prevention, detection and correction of PCR results. Recently, a new family of computational methods for the detection of PCR inhibition called kinetics outlier detection (KOD) emerged. KOD methods are based on comparison of one or a few kinetic parameters describing a test reaction to those describing a set of reference reactions. Modern KOD can detect PCR inhibition reflected by shift of the amplification curve by merely half a cycle with specificity and sensitivity >90%. Based solely on data analysis, these tools complement measures to improve and control pre-analytics. KOD methods do not require labor and materials, do not affect the reaction accuracy and sensitivity and they can be automated for fast and reliable quantification. This review describes the background of KOD methods, their principles, assumptions, strengths and limitations. Finally, the review provides recommendations how to use KOD and how to evaluate its performance.
Figures



References
-
- Murphy Ja, Bustin SA. Reliability of real-time reverse-transcription PCR in clinical diagnostics: gold standard or substandard. Expert. Rev. Mol. Diagn. 2009;9:187–197. - PubMed
-
- International Organization for Standardization. (2005) Microbiology of food and animal feeding stuffs. Polymerase Chain Reaction (PCR) for the detection of food-borne pathogens. General method specific requirements (EN ISO 22174:2005). Geneva.
-
- Lalam NJ. Modelling the PCR amplification process by a size-dependent branching process. Adv. Appl. Probab. 2004;36:602–615.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

