RecA homology search is promoted by mechanical stress along the scanned duplex DNA
- PMID: 22013164
- PMCID: PMC3287184
- DOI: 10.1093/nar/gkr855
RecA homology search is promoted by mechanical stress along the scanned duplex DNA
Abstract
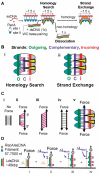
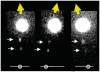
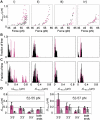
A RecA-single-stranded DNA (RecA-ssDNA) filament searches a genome for sequence homology by rapidly binding and unbinding double-stranded DNA (dsDNA) until homology is found. We demonstrate that pulling on the opposite termini (3' and 5') of one of the two DNA strands in a dsDNA molecule stabilizes the normally unstable binding of that dsDNA to non-homologous RecA-ssDNA filaments, whereas pulling on the two 3', the two 5', or all four termini does not. We propose that the 'outgoing' strand in the dsDNA is extended by strong DNA-protein contacts, whereas the 'complementary' strand is extended by the tension on the base pairs that connect the 'complementary' strand to the 'outgoing' strand. The stress resulting from different levels of tension on its constitutive strands causes rapid dsDNA unbinding unless sufficient homology is present.
Figures



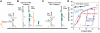
Similar articles
-
Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.Nature. 2020 Oct;586(7831):801-806. doi: 10.1038/s41586-020-2820-9. Epub 2020 Oct 14. Nature. 2020. PMID: 33057191 Free PMC article.
-
Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search.Nature. 2012 Feb 8;482(7385):423-7. doi: 10.1038/nature10782. Nature. 2012. PMID: 22318518 Free PMC article.
-
Complementary strand relocation may play vital roles in RecA-based homology recognition.Nucleic Acids Res. 2012 Nov 1;40(20):10441-51. doi: 10.1093/nar/gks769. Epub 2012 Aug 31. Nucleic Acids Res. 2012. PMID: 22941658 Free PMC article.
-
Structure/function relationships in RecA protein-mediated homology recognition and strand exchange.Crit Rev Biochem Mol Biol. 2015;50(6):453-76. doi: 10.3109/10409238.2015.1092943. Epub 2015 Oct 13. Crit Rev Biochem Mol Biol. 2015. PMID: 26459995 Review.
-
Insights into homology search from cryo-EM structures of RecA-DNA recombination intermediates.Curr Opin Genet Dev. 2021 Dec;71:188-194. doi: 10.1016/j.gde.2021.09.002. Epub 2021 Sep 27. Curr Opin Genet Dev. 2021. PMID: 34592688 Free PMC article. Review.
Cited by
-
Real-time tracking reveals catalytic roles for the two DNA binding sites of Rad51.Nat Commun. 2020 Jun 11;11(1):2950. doi: 10.1038/s41467-020-16750-3. Nat Commun. 2020. PMID: 32528002 Free PMC article.
-
DNA strand exchange and RecA homologs in meiosis.Cold Spring Harb Perspect Biol. 2014 Dec 4;7(1):a016659. doi: 10.1101/cshperspect.a016659. Cold Spring Harb Perspect Biol. 2014. PMID: 25475089 Free PMC article. Review.
-
Tension on dsDNA bound to ssDNA-RecA filaments may play an important role in driving efficient and accurate homology recognition and strand exchange.Phys Rev E Stat Nonlin Soft Matter Phys. 2013 Mar;87(3):032702. doi: 10.1103/PhysRevE.87.032702. Epub 2013 Mar 5. Phys Rev E Stat Nonlin Soft Matter Phys. 2013. PMID: 27499708 Free PMC article.
-
Weaving DNA strands: structural insight on ATP hydrolysis in RecA-induced homologous recombination.Nucleic Acids Res. 2019 Sep 5;47(15):7798-7808. doi: 10.1093/nar/gkz667. Nucleic Acids Res. 2019. PMID: 31372639 Free PMC article.
-
Modeling the Homologous Recombination Process: Methods, Successes and Challenges.Int J Mol Sci. 2023 Oct 4;24(19):14896. doi: 10.3390/ijms241914896. Int J Mol Sci. 2023. PMID: 37834348 Free PMC article. Review.
References
-
- Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange. Annu. Rev. Biochem. 1994;63:991–1043. - PubMed
-
- Roca AI, Cox MM. The RecA protein: structure and function. Crit. Rev. Biochem. Mol. Biol. 1990;25:415–456. - PubMed
-
- Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:17–23. - PubMed
-
- Müller B, Koller T, Stasiak A. Characterization of the DNA binding activity of stable RecA-DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J. Mol. Biol. 1990;212:97–112. - PubMed
-
- Takahashi M, Kubista M, Nordén B. Binding stoichiometry and structure of RecA-DNA complexes studied by flow linear dichroism and fluorescence spectroscopy. Evidence for multiple heterogeneous DNA co-ordination. J. Mol. Biol. 1989;205:137–147. - PubMed

