Direct Quantitative Detection and Identification of Lactococcal Bacteriophages from Milk and Whey by Real-Time PCR: Application for the Detection of Lactococcal Bacteriophages in Goat's Raw Milk Whey in France
- PMID: 22013446
- PMCID: PMC3195528
- DOI: 10.1155/2011/594369
Direct Quantitative Detection and Identification of Lactococcal Bacteriophages from Milk and Whey by Real-Time PCR: Application for the Detection of Lactococcal Bacteriophages in Goat's Raw Milk Whey in France
Abstract
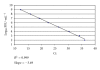
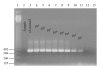
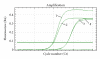
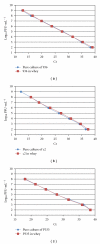
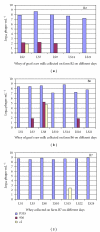
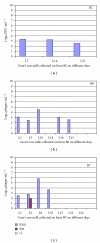
The presence of Lactococcus bacteriophages in milk can partly or completely inhibit milk fermentation. To prevent the problems associated with the bacteriophages, the real-time PCR was developed in this study for direct detection from whey and milk of three main groups of Lactococcus bacteriophages, c2, 936, and P335. The optimization of DNA extraction protocol from complex matrices such as whey and milk was optimized allowed the amplification of PCR without any matrix and nontarget contaminant interference. The real-time PCR program was specific and with the detection limit of 10(2) PFU/mL. The curve slopes were -3.49, -3.69, and -3.45 with the amplification efficiency estimated at 94%, 94%, and 98% and the correlation coefficient (R(2)) of 0.999, 0.999, and 0.998 for c2, 936 and P335 group, respectively. This method was then used to detect the bacteriophages in whey and goat's raw milk coming from three farms located in the Rhône-Alpes region (France).
Figures


References
-
- Franciosi E, Settanni L, Cavazza A, Poznanski E. Biodiversity and technological potential of wild lactic acid bacteria from raw cows’ milk. International Dairy Journal. 2009;19(1):3–11.
-
- Dalmasso M, Hennequin D, Duc C, Demarigny Y. Influence of backslopping on the acidifications curves of “Tomme” type cheeses made during 10 successive days. Journal of Food Engineering. 2009;92(1):50–55.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

