A ssDNA Aptamer That Blocks the Function of the Anti-FLAG M2 Antibody
- PMID: 22013507
- PMCID: PMC3195435
- DOI: 10.4061/2011/720798
A ssDNA Aptamer That Blocks the Function of the Anti-FLAG M2 Antibody
Abstract
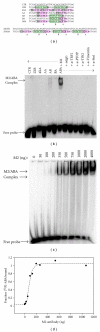
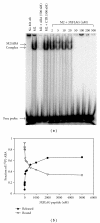
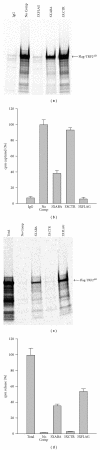
Using SELEX (systematic evolution of ligands by exponential enrichment), we serendipitously discovered a ssDNA aptamer that binds selectively to the anti-FLAG M2 antibody. The aptamer consisted of two motifs (CCTTA and TGTCTWCC) separated by 2-3 bases, and the elimination of one or the other motif abrogated binding. The DNA aptamer and FLAG peptide competed for binding to the antigen-binding pocket of the M2 antibody. In addition, the aptamer eluted FLAG-tagged proteins from the antibody, suggesting a commercial application in protein purification. These findings demonstrate the feasibility of using SELEX to develop ssDNA aptamers that block the function of a specific antibody, a capability that could lead to the development of novel therapeutic modalities for patients with systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases.
Figures

Similar articles
-
An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes.Methods. 2016 Mar 15;97:51-7. doi: 10.1016/j.ymeth.2015.12.005. Epub 2015 Dec 8. Methods. 2016. PMID: 26678795
-
CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for hepatitis C virus envelope glycoprotein E2.PLoS One. 2009 Dec 3;4(12):e8142. doi: 10.1371/journal.pone.0008142. PLoS One. 2009. PMID: 19997645 Free PMC article.
-
Screening and Identifying a Novel ssDNA Aptamer against Alpha-fetoprotein Using CE-SELEX.Sci Rep. 2015 Oct 26;5:15552. doi: 10.1038/srep15552. Sci Rep. 2015. PMID: 26497223 Free PMC article.
-
Development of Cell-Specific Aptamers: Recent Advances and Insight into the Selection Procedures.Molecules. 2017 Nov 27;22(12):2070. doi: 10.3390/molecules22122070. Molecules. 2017. PMID: 29186905 Free PMC article. Review.
-
ABCs of DNA aptamer and related assay development.Biotechnol Adv. 2017 Mar-Apr;35(2):275-301. doi: 10.1016/j.biotechadv.2017.01.003. Epub 2017 Jan 18. Biotechnol Adv. 2017. PMID: 28108354 Review.
Cited by
-
The OB-fold domain 1 of human POT1 recognizes both telomeric and non-telomeric DNA motifs.Biochimie. 2015 Aug;115:17-27. doi: 10.1016/j.biochi.2015.04.015. Epub 2015 Apr 28. Biochimie. 2015. PMID: 25934589 Free PMC article.
-
HOT or not: examining the basis of high-occupancy target regions.Nucleic Acids Res. 2019 Jun 20;47(11):5735-5745. doi: 10.1093/nar/gkz460. Nucleic Acids Res. 2019. PMID: 31114922 Free PMC article.
-
Trends in the Design and Development of Specific Aptamers Against Peptides and Proteins.Protein J. 2016 Apr;35(2):81-99. doi: 10.1007/s10930-016-9653-2. Protein J. 2016. PMID: 26984473 Review.
References
-
- Lyons R, Narain S, Nichols C, Satoh M, Reeves WH. Effective use of autoantibody tests in the diagnosis of systemic autoimmune disease. Annals of the New York Academy of Sciences. 2005;1050:217–228. - PubMed
-
- Pisetsky DS, Caster SA. Binding specificity of a monoclonal anti-DNA antibody. Molecular Immunology. 1982;19(5):645–650. - PubMed
-
- Sasaki T, Muryoi T, Sekiguchi Y, Tamate E, Yoshinaga K, Kitagawa Y. Monoclonal human anti-DNA antibodies from EB virus-transformed lymphocytes of systemic lupus erythematosus (SLE) patients. Journal of Clinical Immunology. 1985;5(4):246–253. - PubMed
-
- DeGiorgio LA, Konstantinov KN, Lee SC, Hardin JA, Volpe BT, Diamond B. A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nature Medicine. 2001;7(11):1189–1193. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

