Synthesis of specifically modified oligonucleotides for application in structural and functional analysis of RNA
- PMID: 22013508
- PMCID: PMC3195551
- DOI: 10.4061/2011/805253
Synthesis of specifically modified oligonucleotides for application in structural and functional analysis of RNA
Abstract
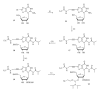
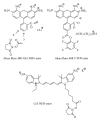
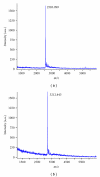
Nowadays, RNA synthesis has become an essential tool not only in the field of molecular biology and medicine, but also in areas like molecular diagnostics and material sciences. Beyond synthetic RNAs for antisense, aptamer, ribozyme, and siRNA technologies, oligoribonucleotides carrying site-specific modifications for structure and function studies are needed. This often requires labeling of the RNA with a suitable spectroscopic reporter group. Herein, we describe the synthesis of functionalized monomer building blocks that upon incorporation in RNA allow for selective reaction with a specific reporter or functional entity. In particular, we report on the synthesis of 5'-O-dimethoxytrityl-2'-O-tert-butyldimethylsilyl protected 3'-O-phosphoramidites of nucleosides that carry amino linkers of different lengths and flexibility at the heterocyclic base, their incorporation in a variety of RNAs, and postsynthetic conjugation with fluorescent dyes and nitroxide spin labels. Further, we show the synthesis of a flavine mononucleotide-N-hydroxy-succinimidyl ester and its conjugation to amino functionalized RNA.
Figures


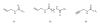








References
-
- Drude I, Dombos V, Vauléon S, Müller S. Drugs made of RNA: development and application of engineered RNAs for gene therapy. Mini-Reviews in Medicinal Chemistry. 2007;7(9):912–931. - PubMed
-
- Meng F, Hennink WE, Zhong Z. Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials. 2009;30(12):2180–2198. - PubMed
-
- Müller S, Wolf J, Ivanov SA. Current strategies for the synthesis of RNA. Current Organic Synthesis. 2004;1(3):293–307.
LinkOut - more resources
Full Text Sources
Other Literature Sources

