Transfer of in vivo primed transgenic T cells supports allergic lung inflammation and FIZZ1 and Ym1 production in an IL-4Rα and STAT6 dependent manner
- PMID: 22014099
- PMCID: PMC3212823
- DOI: 10.1186/1471-2172-12-60
Transfer of in vivo primed transgenic T cells supports allergic lung inflammation and FIZZ1 and Ym1 production in an IL-4Rα and STAT6 dependent manner
Abstract
Background: CD4+ T helper type 2 (TH2) cells, their cytokines IL-4, IL-5 and IL-13 and the transcription factor STAT6 are known to regulate various features of asthma including lung inflammation, mucus production and airway hyperreactivity and also drive alternative activation of macrophages (AAM). However, the precise roles played by the IL-4/IL-13 receptors and STAT6 in inducing AAM protein expression and modulating specific features of airway inflammation are still unclear. Since TH2 differentiation and activation plays a pivotal role in this disease, we explored the possibility of developing an asthma model in mice using T cells that were differentiated in vivo.
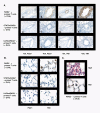
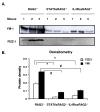
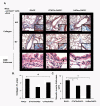
Results: In this study, we monitored the activation and proliferation status of adoptively transferred allergen-specific naïve or in vivo primed CD4+ T cells. We found that both the naïve and in vivo primed T cells expressed similar levels of CD44 and IL-4. However, in vivo primed T cells underwent reduced proliferation in a lymphopenic environment when compared to naïve T cells. We then used these in vivo generated effector T cells in an asthma model. Although there was reduced inflammation in mice lacking IL-4Rα or STAT6, significant amounts of eosinophils were still present in the BAL and lung tissue. Moreover, specific AAM proteins YM1 and FIZZ1 were expressed by epithelial cells, while macrophages expressed only YM1 in RAG2-/- mice. We further show that FIZZ1 and YM1 protein expression in the lung was completely dependent on signaling through the IL-4Rα and STAT6. Consistent with the enhanced inflammation and AAM protein expression, there was a significant increase in collagen deposition and smooth muscle thickening in RAG2-/- mice compared to mice deficient in IL-4Rα or STAT6.
Conclusions: These results establish that transfer of in vivo primed CD4+ T cells can induce allergic lung inflammation. Furthermore, while IL-4/IL-13 signaling through IL-4Rα and STAT6 is essential for AAM protein expression, lung inflammation and eosinophilia are only partially dependent on this pathway. Further studies are required to identify other proteins and signaling pathways involved in airway inflammation.
Figures
Similar articles
-
Absence of the common gamma chain (γ(c)), a critical component of the Type I IL-4 receptor, increases the severity of allergic lung inflammation.PLoS One. 2013 Aug 5;8(8):e71344. doi: 10.1371/journal.pone.0071344. Print 2013. PLoS One. 2013. PMID: 23940740 Free PMC article.
-
Adoptive transfer of IL-4Rα+ macrophages is sufficient to enhance eosinophilic inflammation in a mouse model of allergic lung inflammation.BMC Immunol. 2012 Jan 31;13:6. doi: 10.1186/1471-2172-13-6. BMC Immunol. 2012. PMID: 22292924 Free PMC article.
-
Mechanism of interleukin-25 (IL-17E)-induced pulmonary inflammation and airways hyper-reactivity.Clin Exp Allergy. 2006 Dec;36(12):1575-83. doi: 10.1111/j.1365-2222.2006.02595.x. Clin Exp Allergy. 2006. PMID: 17177681
-
STAT4 and STAT6, their role in cellular and humoral immunity and in diverse human diseases.Int Rev Immunol. 2024;43(6):394-418. doi: 10.1080/08830185.2024.2395274. Epub 2024 Aug 26. Int Rev Immunol. 2024. PMID: 39188021 Review.
-
STAT6 and lung inflammation.JAKSTAT. 2013 Oct 1;2(4):e25301. doi: 10.4161/jkst.25301. Epub 2013 Jun 10. JAKSTAT. 2013. PMID: 24416647 Free PMC article. Review.
Cited by
-
Dissociation of endotoxin tolerance and differentiation of alternatively activated macrophages.J Immunol. 2013 May 1;190(9):4763-72. doi: 10.4049/jimmunol.1202407. Epub 2013 Mar 29. J Immunol. 2013. PMID: 23543762 Free PMC article.
-
An update on Ym1 and its immunoregulatory role in diseases.Front Immunol. 2022 Jul 28;13:891220. doi: 10.3389/fimmu.2022.891220. eCollection 2022. Front Immunol. 2022. PMID: 35967383 Free PMC article. Review.
-
Plexin B1 controls Treg numbers, limits allergic airway inflammation, and regulates mucins.Front Immunol. 2024 Jan 8;14:1297354. doi: 10.3389/fimmu.2023.1297354. eCollection 2023. Front Immunol. 2024. PMID: 38259471 Free PMC article.
-
Mice Expressing Cosegregating Single Nucleotide Polymorphisms (D298G and N397I) in TLR4 Have Enhanced Responses to House Dust Mite Allergen.J Immunol. 2022 May 1;208(9):2085-2097. doi: 10.4049/jimmunol.2100926. Epub 2022 Apr 8. J Immunol. 2022. PMID: 35396219 Free PMC article.
-
Neuroimmune semaphorin 4A as a drug and drug target for asthma.Int Immunopharmacol. 2013 Nov;17(3):568-75. doi: 10.1016/j.intimp.2013.08.005. Epub 2013 Aug 28. Int Immunopharmacol. 2013. PMID: 23994348 Free PMC article.
References
-
- Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

