Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates
- PMID: 22014153
- PMCID: PMC3423228
- DOI: 10.1111/j.1476-5381.2011.01732.x
Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates
Abstract
Background and purpose: Imatinib, a cytochrome P450 2C8 (CYP2C8) and CYP3A4 substrate, markedly increases plasma concentrations of the CYP3A4/5 substrate simvastatin and reduces hepatic CYP3A4/5 activity in humans. Because competitive inhibition of CYP3A4/5 does not explain these in vivo interactions, we investigated the reversible and time-dependent inhibitory effects of imatinib and its main metabolite N-desmethylimatinib on CYP2C8 and CYP3A4/5 in vitro.
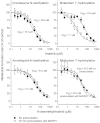
Experimental approach: Amodiaquine N-deethylation and midazolam 1'-hydroxylation were used as marker reactions for CYP2C8 and CYP3A4/5 activity. Direct, IC(50) -shift, and time-dependent inhibition were assessed with human liver microsomes.
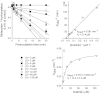
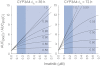
Key results: Inhibition of CYP3A4 activity by imatinib was pre-incubation time-, concentration- and NADPH-dependent, and the time-dependent inactivation variables K(I) and k(inact) were 14.3 µM and 0.072 in(-1) respectively. In direct inhibition experiments, imatinib and N-desmethylimatinib inhibited amodiaquine N-deethylation with a K(i) of 8.4 and 12.8 µM, respectively, and midazolam 1'-hydroxylation with a K(i) of 23.3 and 18.1 µM respectively. The time-dependent inhibition effect of imatinib was predicted to cause up to 90% inhibition of hepatic CYP3A4 activity with clinically relevant imatinib concentrations, whereas the direct inhibition was predicted to be negligible in vivo.
Conclusions and implications: Imatinib is a potent mechanism-based inhibitor of CYP3A4 in vitro and this finding explains the imatinib-simvastatin interaction and suggests that imatinib could markedly increase plasma concentrations of other CYP3A4 substrates. Our results also suggest a possibility of autoinhibition of CYP3A4-mediated imatinib metabolism leading to a less significant role for CYP3A4 in imatinib biotransformation in vivo than previously proposed.
© 2011 The Authors. British Journal of Pharmacology © 2011 The British Pharmacological Society.
Figures
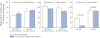

Similar articles
-
Autoinhibition of CYP3A4 leads to important role of CYP2C8 in imatinib metabolism: variability in CYP2C8 activity may alter plasma concentrations and response.Drug Metab Dispos. 2013 Jan;41(1):50-9. doi: 10.1124/dmd.112.048017. Epub 2012 Oct 1. Drug Metab Dispos. 2013. PMID: 23028140
-
Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes.Br J Pharmacol. 2010 Nov;161(5):1059-69. doi: 10.1111/j.1476-5381.2010.00946.x. Br J Pharmacol. 2010. PMID: 20977456 Free PMC article.
-
In vitro assessment of time-dependent inhibitory effects on CYP2C8 and CYP3A activity by fourteen protein kinase inhibitors.Drug Metab Dispos. 2014 Jul;42(7):1202-9. doi: 10.1124/dmd.114.057695. Epub 2014 Apr 8. Drug Metab Dispos. 2014. PMID: 24713129
-
Abiraterone inhibits 1α,25-dihydroxyvitamin D3 metabolism by CYP3A4 in human liver and intestine in vitro.J Steroid Biochem Mol Biol. 2014 Oct;144 Pt A:50-8. doi: 10.1016/j.jsbmb.2013.10.027. Epub 2013 Nov 20. J Steroid Biochem Mol Biol. 2014. PMID: 24269662 Review.
-
Role of Cytochrome P450 2C8 in Drug Metabolism and Interactions.Pharmacol Rev. 2016 Jan;68(1):168-241. doi: 10.1124/pr.115.011411. Pharmacol Rev. 2016. PMID: 26721703 Review.
Cited by
-
CYP2C8 Genotype Significantly Alters Imatinib Metabolism in Chronic Myeloid Leukaemia Patients.Clin Pharmacokinet. 2017 Aug;56(8):977-985. doi: 10.1007/s40262-016-0494-0. Clin Pharmacokinet. 2017. PMID: 27995529
-
Low-dose Imatinib Efficacy in a Gastrointestinal Stromal Tumor Patient With KIT Exon 11 W557_K558 Deletion.In Vivo. 2025 Jan-Feb;39(1):532-538. doi: 10.21873/invivo.13857. In Vivo. 2025. PMID: 39740915 Free PMC article.
-
Clinical Studies on Drug-Drug Interactions Involving Metabolism and Transport: Methodology, Pitfalls, and Interpretation.Clin Pharmacol Ther. 2019 Jun;105(6):1345-1361. doi: 10.1002/cpt.1435. Epub 2019 Apr 20. Clin Pharmacol Ther. 2019. PMID: 30916389 Free PMC article. Review.
-
Molecular Mechanisms of Tyrosine Kinase Inhibitor Resistance in Chronic Myeloid Leukemia.Handb Exp Pharmacol. 2023;280:65-83. doi: 10.1007/164_2023_639. Handb Exp Pharmacol. 2023. PMID: 36882601 Review.
-
Genetic Polymorphisms in CYP2C19 Cause Changes in Plasma Levels and Adverse Reactions to Anlotinib in Chinese Patients With Lung Cancer.Front Pharmacol. 2022 Jun 22;13:918219. doi: 10.3389/fphar.2022.918219. eCollection 2022. Front Pharmacol. 2022. PMID: 35814206 Free PMC article.
References
-
- Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–1029. - PubMed
-
- Bixby D, Talpaz M. Seeking the causes and solutions to imatinib-resistance in chronic myeloid leukemia. Leukemia. 2011;25:7–22. - PubMed
-
- le Coutre P, Kreuzer KA, Pursche S, Bonin M, Leopold T, Baskaynak G, et al. Pharmacokinetics and cellular uptake of imatinib and its main metabolite CGP74588. Cancer Chemother Pharmacol. 2004;53:313–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

