Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer
- PMID: 22014273
- PMCID: PMC3333773
- DOI: 10.1186/gb-2011-12-10-r103
Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer
Abstract
Background: Structural rearrangements form a major class of somatic variation in cancer genomes. Local chromosome shattering, termed chromothripsis, is a mechanism proposed to be the cause of clustered chromosomal rearrangements and was recently described to occur in a small percentage of tumors. The significance of these clusters for tumor development or metastatic spread is largely unclear.
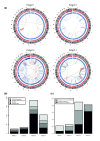
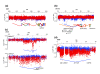
Results: We used genome-wide long mate-pair sequencing and SNP array profiling to reveal that chromothripsis is a widespread phenomenon in primary colorectal cancer and metastases. We find large and small chromothripsis events in nearly every colorectal tumor sample and show that several breakpoints of chromothripsis clusters and isolated rearrangements affect cancer genes, including NOTCH2, EXO1 and MLL3. We complemented the structural variation studies by sequencing the coding regions of a cancer exome in all colorectal tumor samples and found somatic mutations in 24 genes, including APC, KRAS, SMAD4 and PIK3CA. A pairwise comparison of somatic variations in primary and metastatic samples indicated that many chromothripsis clusters, isolated rearrangements and point mutations are exclusively present in either the primary tumor or the metastasis and may affect cancer genes in a lesion-specific manner.
Conclusions: We conclude that chromothripsis is a prevalent mechanism driving structural rearrangements in colorectal cancer and show that a complex interplay between point mutations, simple copy number changes and chromothripsis events drive colorectal tumor development and metastasis.
Figures

Similar articles
-
Prevalence and clinical implications of chromothripsis in cancer genomes.Curr Opin Oncol. 2014 Jan;26(1):64-72. doi: 10.1097/CCO.0000000000000038. Curr Opin Oncol. 2014. PMID: 24305569 Review.
-
An enhanced genetic model of colorectal cancer progression history.Genome Biol. 2019 Aug 15;20(1):168. doi: 10.1186/s13059-019-1782-4. Genome Biol. 2019. PMID: 31416464 Free PMC article.
-
Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline.Hum Mol Genet. 2011 May 15;20(10):1916-24. doi: 10.1093/hmg/ddr073. Epub 2011 Feb 24. Hum Mol Genet. 2011. PMID: 21349919
-
Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms.Genome Res. 2013 May;23(5):762-76. doi: 10.1101/gr.143677.112. Epub 2013 Feb 14. Genome Res. 2013. PMID: 23410887 Free PMC article.
-
The Diverse Effects of Complex Chromosome Rearrangements and Chromothripsis in Cancer Development.Recent Results Cancer Res. 2015;200:165-93. doi: 10.1007/978-3-319-20291-4_8. Recent Results Cancer Res. 2015. PMID: 26376877 Review.
Cited by
-
Chromothripsis and human disease: piecing together the shattering process.Cell. 2012 Jan 20;148(1-2):29-32. doi: 10.1016/j.cell.2012.01.006. Cell. 2012. PMID: 22265399 Free PMC article. Review.
-
The quiescent cellular state is Arf/p53-dependent and associated with H2AX downregulation and genome stability.Int J Mol Sci. 2012;13(5):6492-6506. doi: 10.3390/ijms13056492. Epub 2012 May 24. Int J Mol Sci. 2012. PMID: 22754379 Free PMC article. Review.
-
Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens.BMC Genomics. 2014 Jan 29;15:82. doi: 10.1186/1471-2164-15-82. BMC Genomics. 2014. PMID: 24476156 Free PMC article.
-
Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements.Mol Cell. 2015 Dec 17;60(6):860-72. doi: 10.1016/j.molcel.2015.10.041. Epub 2015 Dec 6. Mol Cell. 2015. PMID: 26669261 Free PMC article.
-
MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes.Anal Chem. 2014 Jul 15;86(14):7150-8. doi: 10.1021/ac502040v. Epub 2014 Jul 3. Anal Chem. 2014. PMID: 24927332 Free PMC article.
References
-
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D. et al.Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. - DOI - PMC - PubMed
-
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. - DOI - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV. et al.The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

