Fast transcription rates of RNA polymerase II in human cells
- PMID: 22015688
- PMCID: PMC3245692
- DOI: 10.1038/embor.2011.196
Fast transcription rates of RNA polymerase II in human cells
Abstract
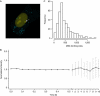
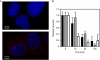
Averaged estimates of RNA polymerase II (RNAPII) elongation rates in mammalian cells have been shown to range between 1.3 and 4.3 kb min(-1). In this work, nascent RNAs from an integrated human immunodeficiency virus type 1-derived vector were detectable at the single living cell level by fluorescent RNA tagging. At steady state, a constant number of RNAs was measured corresponding to a minimal density of polymerases with negligible fluctuations over time. Recovery of fluorescence after photobleaching was complete within seconds, indicating a high rate of RNA biogenesis. The calculated transcription rate above 50 kb min(-1) points towards a wide dynamic range of RNAPII velocities in living cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

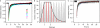
Comment in
-
Pol II caught speeding by single gene imaging.EMBO Rep. 2011 Dec 1;12(12):1208-10. doi: 10.1038/embor.2011.217. EMBO Rep. 2011. PMID: 22094274 Free PMC article. No abstract available.
References
-
- Ardehali MB, Lis JT (2009) Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol 16: 1123–1124 - PubMed
-
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 - PubMed
-
- Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ (2001) Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409: 374–378 - PubMed
-
- Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

