Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading
- PMID: 22016388
- PMCID: PMC3234807
- DOI: 10.1074/jbc.M111.281527
Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading
Abstract
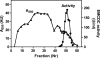
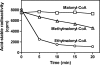
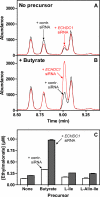
A limited number of enzymes are known that play a role analogous to DNA proofreading by eliminating non-classical metabolites formed by side activities of enzymes of intermediary metabolism. Because few such "metabolite proofreading enzymes" are known, our purpose was to search for an enzyme able to degrade ethylmalonyl-CoA, a potentially toxic metabolite formed at a low rate from butyryl-CoA by acetyl-CoA carboxylase and propionyl-CoA carboxylase, two major enzymes of lipid metabolism. We show that mammalian tissues contain a previously unknown enzyme that decarboxylates ethylmalonyl-CoA and, at lower rates, methylmalonyl-CoA but that does not act on malonyl-CoA. Ethylmalonyl-CoA decarboxylase is particularly abundant in brown adipose tissue, liver, and kidney in mice, and is essentially cytosolic. Because Escherichia coli methylmalonyl-CoA decarboxylase belongs to the family of enoyl-CoA hydratase (ECH), we searched mammalian databases for proteins of uncharacterized function belonging to the ECH family. Combining this database search approach with sequencing data obtained on a partially purified enzyme preparation, we identified ethylmalonyl-CoA decarboxylase as ECHDC1. We confirmed this identification by showing that recombinant mouse ECHDC1 has a substantial ethylmalonyl-CoA decarboxylase activity and a lower methylmalonyl-CoA decarboxylase activity but no malonyl-CoA decarboxylase or enoyl-CoA hydratase activity. Furthermore, ECHDC1-specific siRNAs decreased the ethylmalonyl-CoA decarboxylase activity in human cells and increased the formation of ethylmalonate, most particularly in cells incubated with butyrate. These findings indicate that ethylmalonyl-CoA decarboxylase may correct a side activity of acetyl-CoA carboxylase and suggest that its mutation may be involved in the development of certain forms of ethylmalonic aciduria.
Figures








References
-
- Galperin M. Y., Moroz O. V., Wilson K. S., Murzin A. G. (2006) Mol. Microbiol. 59, 5–19 - PubMed
-
- Van Schaftingen E., Rzem R., Veiga-da-Cunha M. (2009) J. Inherit. Metab. Dis. 32, 135–142 - PubMed
-
- Duran M., Kamerling J. P., Bakker H. D., van Gennip A. H., Wadman S. K. (1980) J. Inherit. Metab. Dis. 3, 109–112 - PubMed
-
- Steenweg M. E., Jakobs C., Errami A., van Dooren S. J., Adeva Bartolomé M. T., Aerssens P., Augoustides-Savvapoulou P., Baric I., Baumann M., Bonafé L., Chabrol B., Clarke J. T., Clayton P., Coker M., Cooper S., Falik-Zaccai T., Gorman M., Hahn A., Hasanoglu A., King M. D., de Klerk H. B., Korman S. H., Lee C., Meldgaard Lund A., Mejaski-Bosnjak V., Pascual-Castroviejo I., Raadhyaksha A., Rootwelt T., Roubertie A., Ruiz-Falco M. L., Scalais E., Schimmel U., Seijo-Martinez M., Suri M., Sykut-Cegielska J., Trefz F. K., Uziel G., Valayannopoulos V., Vianey-Saban C., Vlaho S., Vodopiutz J., Wajner M., Walter J., Walter-Derbort C., Yapici Z., Zafeiriou D. I., Spreeuwenberg M. D., Celli J., den Dunnen J. T., van der Knaap M. S., Salomons G. S. (2010) Hum. Mutat. 31, 380–390 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

