Protein interactions and localization of the Escherichia coli accessory protein HypA during nickel insertion to [NiFe] hydrogenase
- PMID: 22016389
- PMCID: PMC3234881
- DOI: 10.1074/jbc.M111.290726
Protein interactions and localization of the Escherichia coli accessory protein HypA during nickel insertion to [NiFe] hydrogenase
Abstract
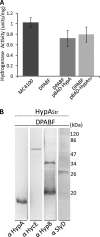
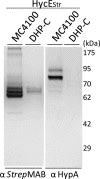
Nickel delivery during maturation of Escherichia coli [NiFe] hydrogenase 3 includes the accessory proteins HypA, HypB, and SlyD. Although the isolated proteins have been characterized, little is known about how they interact with each other and the hydrogenase 3 large subunit, HycE. In this study the complexes of HypA and HycE were investigated after modification with the Strep-tag II. Multiprotein complexes containing HypA, HypB, SlyD, and HycE were observed, consistent with the assembly of a single nickel insertion cluster. An interaction between HypA and HycE did not require the other nickel insertion proteins, but HypB was not found with the large subunit in the absence of HypA. The HypA-HycE complex was not detected in the absence of the HypC or HypD proteins, involved in the preceding iron insertion step, and this interaction is enhanced by nickel brought into the cell by the NikABCDE membrane transporter. Furthermore, without the hydrogenase 1, 2, and 3 large subunits, complexes between HypA, HypB, and SlyD were observed. These results support the hypothesis that HypA acts as a scaffold for assembly of the nickel insertion proteins with the hydrogenase precursor protein after delivery of the iron center. At different stages of the hydrogenase maturation process, HypA was observed at or near the cell membrane by using fluorescence confocal microscopy, as was HycE, suggesting membrane localization of the nickel insertion event.
Figures

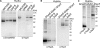
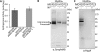


Similar articles
-
The Escherichia coli metal-binding chaperone SlyD interacts with the large subunit of [NiFe]-hydrogenase 3.FEBS Lett. 2011 Jan 21;585(2):291-4. doi: 10.1016/j.febslet.2010.12.024. Epub 2010 Dec 23. FEBS Lett. 2011. PMID: 21185288
-
The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD.J Biol Chem. 2007 Jun 1;282(22):16177-86. doi: 10.1074/jbc.M610834200. Epub 2007 Apr 10. J Biol Chem. 2007. PMID: 17426034
-
Complex formation between the Escherichia coli [NiFe]-hydrogenase nickel maturation factors.Biometals. 2019 Jun;32(3):521-532. doi: 10.1007/s10534-019-00173-9. Epub 2019 Feb 13. Biometals. 2019. PMID: 30758762
-
Structural Insight into [NiFe] Hydrogenase Maturation by Transient Complexes between Hyp Proteins.Acc Chem Res. 2020 Apr 21;53(4):875-886. doi: 10.1021/acs.accounts.0c00022. Epub 2020 Mar 31. Acc Chem Res. 2020. PMID: 32227866 Review.
-
Moving nickel along the hydrogenase-urease maturation pathway.Metallomics. 2022 May 13;14(5):mfac003. doi: 10.1093/mtomcs/mfac003. Metallomics. 2022. PMID: 35556134 Review.
Cited by
-
Comparative genomic analysis of nickel homeostasis in cable bacteria.BMC Genomics. 2024 Jul 15;25(1):692. doi: 10.1186/s12864-024-10594-7. BMC Genomics. 2024. PMID: 39009997 Free PMC article.
-
Transition Metal Transport in Plants and Associated Endosymbionts: Arbuscular Mycorrhizal Fungi and Rhizobia.Front Plant Sci. 2016 Jul 29;7:1088. doi: 10.3389/fpls.2016.01088. eCollection 2016. Front Plant Sci. 2016. PMID: 27524990 Free PMC article. Review.
-
High-affinity metal binding by the Escherichia coli [NiFe]-hydrogenase accessory protein HypB is selectively modulated by SlyD.Metallomics. 2017 May 24;9(5):482-493. doi: 10.1039/c7mt00037e. Metallomics. 2017. PMID: 28352890 Free PMC article.
-
Selective inhibition of NikA mediated Ni(II) import in E. coli by the Indium(III)-EDTA complex.Metallomics. 2025 Mar 28;17(4):mfaf008. doi: 10.1093/mtomcs/mfaf008. Metallomics. 2025. PMID: 40037903 Free PMC article.
-
Structural investigations connect the disordered N-terminal extension of HypB to the activities of HypB and SlyD in E. coli.Protein Sci. 2025 Aug;34(8):e70231. doi: 10.1002/pro.70231. Protein Sci. 2025. PMID: 40698646 Free PMC article.
References
-
- Forzi L., Sawers R. G. (2007) Biometals 20, 565–578 - PubMed
-
- Redwood M. D., Mikheenko I. P., Sargent F., Macaskie L. E. (2008) FEMS Microbiol. Lett. 278, 48–55 - PubMed
-
- McGlynn S. E., Mulder D. W., Shepard E. M., Broderick J. B., Peters J. W. (2009) Dalton Trans 22, 4274–4285 - PubMed
-
- Fontecilla-Camps J. C., Volbeda A., Cavazza C., Nicolet Y. (2007) Chem. Rev. 107, 4273–4303 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

