Misalignment of PLP/DM20 transmembrane domains determines protein misfolding in Pelizaeus-Merzbacher disease
- PMID: 22016529
- PMCID: PMC6623585
- DOI: 10.1523/JNEUROSCI.2097-11.2011
Misalignment of PLP/DM20 transmembrane domains determines protein misfolding in Pelizaeus-Merzbacher disease
Abstract
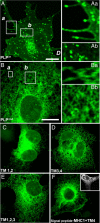
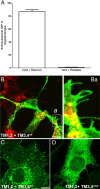
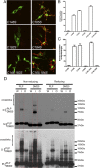
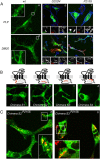
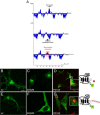
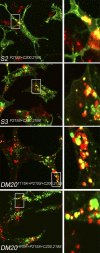
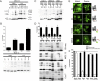
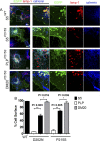
A large number of genetic diseases have been associated with truncated or misfolded membrane proteins trapped in the endoplasmic reticulum (ER). In the ER, they activate the unfolded protein response, which can trigger cell death. Hence, a better understanding of protein misfolding features might help in developing novel therapies. Here, we have studied the molecular basis of Pelizaeus-Merzbacher disease, a leukodystrophy defined by mutations of the PLP1 gene and ER retention of two encoded tetraspan myelin proteins, PLP and DM20. In mouse oligodendroglial cells, mutant isoforms of PLP/DM20 with fewer than all four transmembrane (TM) domains are fully ER retained. Surprisingly, a truncated PLP with only two N-terminal TM domains shows normal cell-surface expression when coexpressed with a second truncated PLP harboring the two C-terminal TM domains. This striking ability to properly self-align the TM domains is disease relevant, as shown for the smaller splice isoform DM20. Here, the increased length of TM domain 3 allows for compensation of the effect of several PLP1 point mutations that impose a conformational constraint onto the adjacent extracellular loop region. We conclude that an important determinant in the quality control of polytopic membrane proteins is the free alignment of their TM domains.
Figures

References
-
- Albrecht C, Baynes K, Sardini A, Schepelmann S, Eden ER, Davies SW, Higgins CF, Feher MD, Owen JS, Soutar AK. Two novel missense mutations in ABCA1 result in altered trafficking and cause severe autosomal recessive HDL deficiency. Biochim Biophys Acta. 2004;1689:47–57. - PubMed
-
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
