CAFET algorithm reveals Wnt/PCP signature in lung squamous cell carcinoma
- PMID: 22016777
- PMCID: PMC3189939
- DOI: 10.1371/journal.pone.0025807
CAFET algorithm reveals Wnt/PCP signature in lung squamous cell carcinoma
Abstract
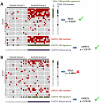
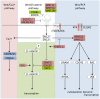
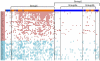
We analyzed the gene expression patterns of 138 Non-Small Cell Lung Cancer (NSCLC) samples and developed a new algorithm called Coverage Analysis with Fisher's Exact Test (CAFET) to identify molecular pathways that are differentially activated in squamous cell carcinoma (SCC) and adenocarcinoma (AC) subtypes. Analysis of the lung cancer samples demonstrated hierarchical clustering according to the histological subtype and revealed a strong enrichment for the Wnt signaling pathway components in the cluster consisting predominantly of SCC samples. The specific gene expression pattern observed correlated with enhanced activation of the Wnt Planar Cell Polarity (PCP) pathway and inhibition of the canonical Wnt signaling branch. Further real time RT-PCR follow-up with additional primary tumor samples and lung cancer cell lines confirmed enrichment of Wnt/PCP pathway associated genes in the SCC subtype. Dysregulation of the canonical Wnt pathway, characterized by increased levels of β-catenin and epigenetic silencing of negative regulators, has been reported in adenocarcinoma of the lung. Our results suggest that SCC and AC utilize different branches of the Wnt pathway during oncogenesis.
Conflict of interest statement
Figures
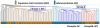

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

