Acylpeptide hydrolase inhibition as targeted strategy to induce proteasomal down-regulation
- PMID: 22016782
- PMCID: PMC3189933
- DOI: 10.1371/journal.pone.0025888
Acylpeptide hydrolase inhibition as targeted strategy to induce proteasomal down-regulation
Abstract
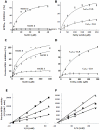
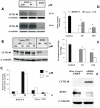
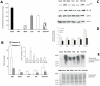
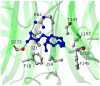
Acylpeptide hydrolase (APEH), one of the four members of the prolyl oligopeptidase class, catalyses the removal of N-acylated amino acids from acetylated peptides and it has been postulated to play a key role in protein degradation machinery. Disruption of protein turnover has been established as an effective strategy to down-regulate the ubiquitin-proteasome system (UPS) and as a promising approach in anticancer therapy.Here, we illustrate a new pathway modulating UPS and proteasome activity through inhibition of APEH. To find novel molecules able to down-regulate APEH activity, we screened a set of synthetic peptides, reproducing the reactive-site loop of a known archaeal inhibitor of APEH (SsCEI), and the conjugated linoleic acid (CLA) isomers. A 12-mer SsCEI peptide and the trans10-cis12 isomer of CLA, were identified as specific APEH inhibitors and their effects on cell-based assays were paralleled by a dose-dependent reduction of proteasome activity and the activation of the pro-apoptotic caspase cascade. Moreover, cell treatment with the individual compounds increased the cytoplasm levels of several classic hallmarks of proteasome inhibition, such as NFkappaB, p21, and misfolded or polyubiquitinylated proteins, and additive effects were observed in cells exposed to a combination of both inhibitors without any cytotoxicity. Remarkably, transfection of human bronchial epithelial cells with APEH siRNA, promoted a marked accumulation of a mutant of the cystic fibrosis transmembrane conductance regulator (CFTR), herein used as a model of misfolded protein typically degraded by UPS. Finally, molecular modeling studies, to gain insights into the APEH inhibition by the trans10-cis12 CLA isomer, were performed.Our study supports a previously unrecognized role of APEH as a negative effector of proteasome activity by an unknown mechanism and opens new perspectives for the development of strategies aimed at modulation of cancer progression.
Conflict of interest statement
Figures


References
-
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. - PubMed
-
- Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. - PubMed
-
- Shah IM, Di Napoli M. The ubiquitin-proteasome system and proteasome inhibitors in central nervous system diseases. Cardiovasc Hematol Disord Drug Targets. 2007;7:250–273. - PubMed
-
- Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J Clin Oncol. 2005;23:630–639. - PubMed
-
- Naujokat C, Sezer O, Zinke H, Leclere A, Hauptmann S, et al. Proteasome inhibitors induced caspase-dependent apoptosis and accumulation of p21WAF1/Cip1 in human immature leukemic cells. Eur J Haematol. 2000;65:221–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

