Catalytic properties of the isolated diaphorase fragment of the NAD-reducing [NiFe]-hydrogenase from Ralstonia eutropha
- PMID: 22016788
- PMCID: PMC3189943
- DOI: 10.1371/journal.pone.0025939
Catalytic properties of the isolated diaphorase fragment of the NAD-reducing [NiFe]-hydrogenase from Ralstonia eutropha
Abstract
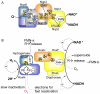
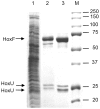
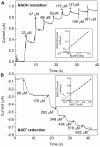
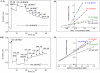
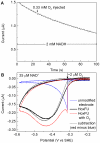
The NAD+-reducing soluble hydrogenase (SH) from Ralstonia eutropha H16 catalyzes the H₂-driven reduction of NAD+, as well as reverse electron transfer from NADH to H+, in the presence of O₂. It comprises six subunits, HoxHYFUI₂, and incorporates a [NiFe] H+/H₂ cycling catalytic centre, two non-covalently bound flavin mononucleotide (FMN) groups and an iron-sulfur cluster relay for electron transfer. This study provides the first characterization of the diaphorase sub-complex made up of HoxF and HoxU. Sequence comparisons with the closely related peripheral subunits of Complex I in combination with UV/Vis spectroscopy and the quantification of the metal and FMN content revealed that HoxFU accommodates a [2Fe2S] cluster, FMN and a series of [4Fe4S] clusters. Protein film electrochemistry (PFE) experiments show clear electrocatalytic activity for both NAD+ reduction and NADH oxidation with minimal overpotential relative to the potential of the NAD+/NADH couple. Michaelis-Menten constants of 56 µM and 197 µM were determined for NADH and NAD+, respectively. Catalysis in both directions is product inhibited with K(I) values of around 0.2 mM. In PFE experiments, the electrocatalytic current was unaffected by O₂, however in aerobic solution assays, a moderate superoxide production rate of 54 nmol per mg of protein was observed, meaning that the formation of reactive oxygen species (ROS) observed for the native SH can be attributed mainly to HoxFU. The results are discussed in terms of their implications for aerobic functioning of the SH and possible control mechanism for the direction of catalysis.
Conflict of interest statement
Figures



References
-
- Friedrich T, Scheide D. The respiratory Complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5. - PubMed
-
- Albracht SPJ, van der Linden E, Faber BW. Quantitative amino acid analysis of bovine NADH:ubiquinone oxidoreductase (Complex I) and related enzymes. Consequences for the number of prosthetic groups. Biochim Biophys Acta. 2003;1557:41–49. - PubMed
-
- Cramm R. Genomic view of energy metabolism in Ralstonia eutropha H16. J Mol Microbiol Biotechnol. 2009;16:38–52. - PubMed
-
- Burgdorf T, Lenz O, Buhrke T, van der Linden E, Jones AK, et al. [NiFe]-hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen-tolerant biological hydrogen oxidation. J Mol Microbiol Biotechnol. 2005;10:181–196. - PubMed
-
- Burgdorf T, Löscher S, Liebisch P, van der Linden E, Galander M, et al. Structural and oxidation-state changes at its nonstandard Ni-Fe site during activation of the NAD-reducing hydrogenase from Ralstonia eutropha detected by X-ray absorption, EPR, and FTIR spectroscopy. J Am Chem Soc. 2005;127:576–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

