Adapting to life at the end of the line: How Drosophila telomeric retrotransposons cope with their job
- PMID: 22016861
- PMCID: PMC3190324
- DOI: 10.4161/mge.1.2.16914
Adapting to life at the end of the line: How Drosophila telomeric retrotransposons cope with their job
Abstract
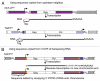
Drosophila telomeres are remarkable because they are maintained by telomere-specific retrotransposons, rather than the enzyme telomerase that maintains telomeres in almost every other eukaryotic organism. Successive transpositions of the Drosophila retrotransposons onto chromosome ends produce long head-to-tail arrays that are analogous in form and function to the long arrays of short repeats produced by telomerase in other organisms. Nevertheless, Drosophila telomere repeats are retrotransposons, complex entities three orders of magnitude longer than simple telomerase repeats. During the >40-60 My they have been coevolving with their host, these retrotransposons perforce have evolved a complex relationship with Drosophila cells to maintain populations of active elements while carrying out functions analogous to those of telomerase repeats in other organisms. Although they have assumed a vital role in maintaining the Drosophila genome, the three Drosophila telomere-specific elements are non-LTR retrotransposons, closely related to some of the best known non-telomeric elements in the Drosophila genome. Thus, these elements offer an opportunity to study ways in which retrotransposons and their host cells can coevolve cooperatively. The telomere-specific elements display several characteristics that appear important to their roles at the telomere; for example, we have recently reported that they have evolved at least two innovative mechanisms for protecting essential sequence on their 5'ends. Because every element serves as the end of the chromosome immediately after it transposes, its 5'end is subject to chromosomal erosion until it is capped by a new transposition. These two mechanisms make it possible for at least a significant fraction of elements to survive their initial time as the chromosome end without losing sequence necessary to be competent for subsequent transposition. Analysis of sequence from >90 kb of assembled telomere array shows that these mechanisms for small scale sequence protection are part of a unified set which maintains telomere length homeostasis. Here we concentrate on recently elucidated mechanisms that have evolved to provide this small scale 5' protection.
Figures

References
-
- Muller HJ. The remaking of chromosomes. Collecting Net. 1938;13:181–195.
-
- Blackburn EH. Telomerases. Annu Rev Biochem. 1992;61:113–129. - PubMed
-
- Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. - PubMed
-
- Martinez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
