How do pleiotropic kinase hubs mediate specific signaling by TNFR superfamily members?
- PMID: 22017429
- PMCID: PMC3357464
- DOI: 10.1111/j.1600-065X.2011.01060.x
How do pleiotropic kinase hubs mediate specific signaling by TNFR superfamily members?
Abstract
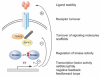
Tumor necrosis factor receptor (TNFR) superfamily members mediate the cellular response to a wide variety of biological inputs. The responses range from cell death, survival, differentiation, proliferation, to the regulation of immunity. All these physiological responses are regulated by a limited number of highly pleiotropic kinases. The fact that the same signaling molecules are involved in transducing signals from TNFR superfamily members that regulate different and even opposing processes raises the question of how their specificity is determined. Regulatory strategies that can contribute to signaling specificity include scaffolding to control kinase specificity, combinatorial use of several signal transducers, and temporal control of signaling. In this review, we discuss these strategies in the context of TNFR superfamily member signaling.
© 2011 John Wiley & Sons A/S.
Figures

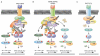


References
-
- Aggarwal BB. Signaling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. - PubMed
-
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. - PubMed
-
- Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. - PubMed
-
- Vandenabeele P, Declercq W, Van Herreweghe F, Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal. 2010;3:re4. - PubMed
-
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

