NMR studies of localized water and protein backbone dynamics in mechanically strained elastin
- PMID: 22017547
- PMCID: PMC3622950
- DOI: 10.1021/jp207607r
NMR studies of localized water and protein backbone dynamics in mechanically strained elastin
Abstract
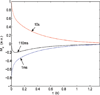
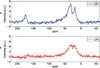
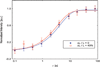
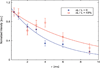
We report on measurements of the dynamics of localized waters of hydration and the protein backbone of elastin, a remarkable resilient protein found in vertebrate tissues, as a function of the applied external strain. Using deuterium 2D T(1)-T(2) NMR, we separate four reservoirs in the elastin-water system characterized by water with distinguishable mobilities. The measured correlation times corresponding to random tumbling of water localized to the protein is observed to decrease with increasing strain and is interpreted as an increase in its orientational entropy. The NMR T(1) and T(1ρ) relaxation times of the carbonyl and aliphatic carbons of the protein backbone are measured and indicate a reduction in the correlation time as the elastomer strain is increased. It is argued, and supported by MD simulation of a short model elastin peptide [VPGVG](3), that the observed changes in the backbone dynamics give rise to the development of an entropic elastomeric force that is responsible for elastins' remarkable elasticity.
© 2011 American Chemical Society
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

